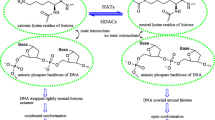
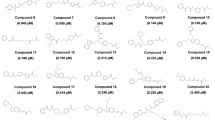
Abstract
Histone deacetylases (HDACs) were highlighted as a novel category of anticancer targets. Several HDACs inhibitors were approved for therapeutic use in cancer treatment. Comparatively, receptor-dependent 4D-QSAR, LQTA-QSAR, is a new approach which generates conformational ensemble profiles of compounds by molecular dynamics simulations at binding site of enzyme. This work describes a receptor-dependent 4D-QSAR studies on hydroxamate-based HDACs inhibitors. The 4D-QSAR model was generated by multiple linear regression method of QSARINS. Leave-N-out cross-validation (LNO) and Y-randomization were performed to analysis of the independent test set and to verify the robustness of the model. Best 4D-QSAR model showed the following statistics: R2 = 0.8117, Q2LOO = 0.6881, Q2LNO = 0.6830, R2Pred = 0.884. The results may be used for further virtual screening and design for novel HDACs inhibitors.
Graphical abstract
The receptor dependent 4D-QSAR model was developed for the hydroxamate derivatives as HDAC inhibitors by making use of molecular dynamics simulation to obtain conformational ensemble profile for each compound. The multiple linear regression method was used to generate 4D-QSAR model with the suitable predictive ability and the excellent statistical parameters.









Similar content being viewed by others
References
Krämer OH, Göttlicher M, Heinzel T (2001) Histone deacetylase as a therapeutic target. Trends Endocrinol Metab 12(7):294–300. https://doi.org/10.1016/s1043-2760(01)00438-6
Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK (2001) Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer 1(3):194–202. https://doi.org/10.1038/35106079
Grozinger CM, Schreiber SL (2002) Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem Biol 9(1):3–16. https://doi.org/10.1016/s1074-5521(02)00092-3
Johnstone RW (2002) Histone-deacetylase inhibitors: novel drugs for the treatment of cancer. Nat Rev Drug Discov 1(4):287–299. https://doi.org/10.1038/nrd772
Marks PA, Breslow R (2007) Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol 25(1):84–90. https://doi.org/10.1038/nbt1272
Marshall JL, Rizvi N, Kauh J, Dahut W, Figuera M, Kang MH, Figg WD, Wainer I, Chaissang C, Li MZ, Hawkins MJ (2002) A phase I trial of depsipeptide (FR901228) in patients with advanced cancer. J Exp Ther Oncol 2(6):325–332. https://doi.org/10.1046/j.1359-4117.2002.01039.x
Shim J, Mackerell AD (2011) Computational ligand-based rational design: role of conformational sampling and force fields in model development. Medchemcomm 2(5):356–370. https://doi.org/10.1039/C1MD00044F
Ghasemi JB, Salahinejad M, Rofouei MK (2011) Review of the quantitative structure–activity relationship modelling methods on estimation of formation constants of macrocyclic compounds with different guest molecules. Supramol Chem 23(9):614–629. https://doi.org/10.1080/10610278.2011.581281
Hopfinger AJ, Wang S, Tokarski JS, Jin B, Albuquerque MG, Madhav JP, Duraiswami C (1997) Construction of 3D-QSAR models using the 4D-QSAR analysis formalism. J Am Chem Soc 119(43):10509–10524. https://doi.org/10.1021/ja9718937
Martins JPA, Barbosa EG, Pasqualoto KFM, Ferreira MMC (2009) LQTA-QSAR: a new 4D-QSAR methodology. J Chem Inf Model 49(6):1428–1436. https://doi.org/10.1021/ci900014f
Santosfilho OA, Hopfinger AJ (2002) The 4D-QSAR paradigm: application to a novel set of non- peptidic HIV protease inhibitors. Quant Struct-Act Relat 21(4):369–381. https://doi.org/10.1002/1521-3838(200210)21:4%3c369::AID-QSAR369%3e3.0.CO;2-1
Dassault SystemesBIOVIA, Discovery Studio, San Diego, California, USA. http://accelrys.com/
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and Autodocktools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791. https://doi.org/10.1002/jcc.21256
Csizmadia P, MarvinSketch and MarvinView: molecule applets for the world wide web, https://chemaxon.com/products/marvin
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera–a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612. https://doi.org/10.1002/jcc.20084
Thompson MA, Planaria Software LLC, ArgusLab-molecular modeling, graphics & drug design program. http://www.arguslab.com/
DeLano WL (2002) The PyMOL molecular graphics system. http://www.pymol.org.
Pronk S, Pll S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, Shirts MR, Smith JC, Kasson PM, Van Der Spoel D, Hess B, Lindahl E (2013) a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29(7):845–854. https://doi.org/10.1093/bioinformatics/btt055
Duke R, Giese T, Gohlke H (2016) AmberTools. University of California, San Francisco. http://ambermd.org/AmberTools.php
Ray BD (2010) GROMACS Contribution Section http://www.gromacs.org/Downloads/User_contributions/Other_software
Gramatica P, Chirico N, Papa E, Cassani S, Kovarich S (2013) QSARINS: a new software for the development, analysis, and validation of QSAR MLR models. J Comput Chem 34(24):2121–2132. https://doi.org/10.1002/jcc.23361
Lavoie R, Bouchain G, Frechette S, Woo SH, Abou-Khalil E, Leit S, Fournel M, Yan PT, Trachy-Bourget MC, Beaulieu C, Li Z, Besterman J, Delorme D (2001) Design and synthesis of a novel class of histone deacetylase inhibitors. Bioorg Med Chem Lett 11(21):2847–2850. https://doi.org/10.1016/s0960-894x(01)00552-2
Remiszewski SW, Sambucetti LC, Atadja P, Bair KW, Cornell WD, Green MA, Howell KL, Jung M, Kwon P, Trogani N, Walker H (2002) Inhibitors of human histone deacetylase: synthesis and enzyme and cellular activity of straight chain hydroxamates. J Med Chem 45(4):753–757. https://doi.org/10.1021/jm015568c
Woo SH, Frechette S, Abou Khalil E, Bouchain G, Vaisburg A, Bernstein N, Moradei O, Leit S, Allan M, Fournel M, Trachy-Bourget M-C, Li Z, Besterman JM, Delorme D (2002) Structurally simple trichostatin a-like straight chain hydroxamates as potent histone deacetylase inhibitors. J Med Chem 45(13):2877–2885. https://doi.org/10.1021/jm020154k
Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP (1999) Structures of a histone deacetylase homologue bound to the tsa and saha inhibitors. Nature 401(6749):188–193. https://doi.org/10.1038/43710
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pkas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371. https://doi.org/10.1093/nar/gki464
Mark P, Nilsson L (2001) Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J Phys Chem A 105(43):9954–9960. https://doi.org/10.1021/jp003020w
Darden T, York D, Pedersen L (1993) Particle mesh ewald: an N-Log(N) method for ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
MacKerell Jr AD, Banavali NK (2000) All‐atom empirical force field for nucleic acids: II. application to molecular dynamics simulations of DNA and RNA in solution. J Comput Chem 21(2): 105–120. https://doi.org/https://doi.org/10.1002/(SICI)1096-987X(20000130)21:2<105::AID-JCC3>3.0.CO;2-P
Barbosa EG, Pasqualoto KFM, Ferreira MMC (2012) The receptor-dependent LQTA-QSAR: application to a set of trypanothione reductase inhibitors. J Comput Aided Mol Des 26(9):1055–1065. https://doi.org/10.1007/s10822-012-9598-2
Patil RB, Barbosa EG, Sangshetti JN, Sawant SD, Zambre VP (2018) LQTA-R: a new 3D-QSAR methodology applied to a set of DGAT1 inhibitors. Comput Biol Chem 74:123–131. https://doi.org/10.1016/j.compbiolchem.2018.02.021
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20(4):269–276. https://doi.org/10.1016/s1093-3263(01)00123-1
Golbraikh A, Tropsha A (2002) Predictive QSAR modeling based on diversity sampling of experimental datasets for the training and test set selection. J Comput Aided Mol Des 16(5–6):357–369. https://doi.org/10.1023/a:1020869118689
Chirico N, Gramatica P (2011) Real external predictivity of QSAR models: how to evaluate it? comparison of different validation criteria and proposal of using the concordance correlation coefficient. J Chem Inf Model 51(9):2320–2335. https://doi.org/10.1021/ci200211n
Ghasemi JB, Safavi-Sohi R, Barbosa EG (2012) 4D-LQTA-QSAR and docking study on potent gram-negative specific LPXC inhibitors: a comparison to COMFA modeling. Mol Divers 16(1):203–213. https://doi.org/10.1007/s11030-011-9340-3
Acknowledgements
The project was supported by the Nanchang University Teaching Reform Foundation (NCUJGLX-19-130, NCUJGLX-19-124), the Graduate Innovation Foundation of Jiangxi Province (CX2018190) and the Undergraduate Innovation and Entrepreneurship Foundation (2020CX289)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares no conflict of interest, financial or otherwise.
Human and animal rights
No Animals/Humans were used for studies that are basis of this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the Supplementary Information.
Rights and permissions
About this article
Cite this article
Hu, Z., Lin, Q., Liu, H. et al. Molecular dynamics-guided receptor-dependent 4D-QSAR studies of HDACs inhibitors. Mol Divers 26, 757–768 (2022). https://doi.org/10.1007/s11030-021-10181-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10181-y