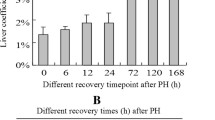
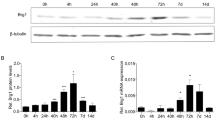
Abstract
The ubiquitin signalling pathway is a large system associated with numerous intracellular mechanisms. However, its function in the liver regeneration process remains unknown. This particular study investigates the intracellular effect mechanisms of the ubiquitin signalling pathway. It also determines the differences in the expression of 88 genes belonging to the ubiquitin pathway using the RT-PCR array method. To conduct this research, three genes—that differed in the expression analysis were selected. Moreover, their proteins were analysed by western blot analysis while using Ki67 immunohistochemical analysis that determines proliferation rates by hour. It was determined that BRCA1 and BARD1, which are effective in DNA repair, play an active role at PH24. Similarly, Ube2t expression, which belongs to the Fanconi anaemia pathway associated with DNA repair, was also found to be high at PH12-72 h. In addition, it was revealed that the expressions of Anapc2, Anapc11, Cdc20 belonging to the APC/CCdc20 complex, which participate in cell cycle regulation, differed at different hours after PH. Expression of Mul1, which is involved in mitochondrial biogenesis and mitophagy mechanisms, peaked at PH12 under the observation. Considering the Mul1 gene expression difference, MUL1-mediated mitophagy and mitochondrial fission mechanism may be associated with liver regeneration. It was also determined that PARKIN-mediated mitophagy mechanisms are not active in 0–72 h of liver regeneration since PARKIN expression did not show a significant change in PH groups.







Similar content being viewed by others
Data availability
The online version of RT-PCR Array data (unnormalized/normalised/fold change) available at: https://github.com/aaozmen/aaozmen
References
Hu W, Nevzorova YA, Haas U et al (2014) Concurrent deletion of cyclin E1 and cyclin-dependent kinase 2 in hepatocytes inhibits DNA replication and liver regeneration in mice. Hepatology 59(2):651–660. https://doi.org/10.1002/hep.26584
Taub R (2004) Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Bio 5(10):836–847. https://doi.org/10.1038/nrm1489
Fausto N (2000) Liver regeneration. J Hepatol 32:19–31. https://doi.org/10.1016/S0168-8278(00)80412-2
Michalopoulos GK (2010) Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 176(1):2–13. https://doi.org/10.2353/ajpath.2010.090675
Abu Rmilah A, Zhou W, Nelson E et al (2019) Understanding the marvels behind liver regeneration. WIREs Dev Biol 8(e340):1–28. https://doi.org/10.1002/wdev.340
Xu C, Chang C, Yuan J et al (2005) Expressed genes in regenerating rat liver after partial hepatectomy. World J Gastroenterol 11(19):2932–2940. https://doi.org/10.3748/wjg.v11.i19.2932
Wang X, Herr RA, Rabelink M et al (2009) Ube2j2 ubiquitinates hydroxylated amino acids on ER-associated degradation substrates. J Cell Biol 187(5):655–668. https://doi.org/10.1083/jcb.200908036
Kurinna S, Barton MC (2011) Cascades of transcription regulation during liver regeneration. Int J Biochem Cell B 43(2):189–197. https://doi.org/10.1016/j.biocel.2010.03.013
Ciechanover A (1998) The ubiquitin—proteasome pathway: on protein death and cell life. EMBO J 17(24):7151–7160. https://doi.org/10.1093/emboj/17.24.7151
Yau R, Rape M (2016) The increasing complexity of the ubiquitin code. Nat Cell Biol 18(6):579–586. https://doi.org/10.1038/ncb3358
Zinngrebe J, Montinaro A, Peltzer N et al (2013) Ubiquitin in the immune system. EMBO Rep 15(1):28–45. https://doi.org/10.1002/embr.201338025
Koepp DM, Harper JW, Elledge SJ (1999) How the cyclin became a cyclin: regulated proteolysis in the cell cycle. Cell 97:431–434. https://doi.org/10.1016/s0092-8674(00)80753-9
Peters JM (2002) The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol Cell 9:931–943. https://doi.org/10.1016/s1097-2765(02)00540-3
Wu SY, Kuan VJW, Tzeng YW et al (2016) The anaphase promoting complex works together with the SCF complex for proteolysis of the S-phase cyclin Clb6 during the transition from G1 to S phase. Fungal Genet Biol 91(2016):6–19. https://doi.org/10.1016/j.fgb.2016.03.004
Garcia-Higuera I, Taniguchi T, Ganesan S et al (2001) Interaction of the Fanconi Anemia proteins and BRCA1 in a common pathway. Mol Cell 7(2):249–262. https://doi.org/10.1016/S1097-2765(01)00173-3
Cheung RS, Taniguchi T (2017) Recent insights into the molecular basis of Fanconi anemia: genes, modifiers, and drivers. Int J Hematol 106(3):335–344. https://doi.org/10.1007/s12185-017-2283-4
Deng CX (2002) Roles of BRCA1 in centrosome duplication. Oncogene 21:6222–6227. https://doi.org/10.1038/sj.onc.1205713
Kais Z, Parvin JD (2008) Regulation of centrosomes by the BRCA1-dependent ubiquitin ligase. Cancer Biol Ther 7(10):1540–1543. https://doi.org/10.4161/cbt.7.10.7053
Villa E, Marchetti S, Ricci JE (2018) No Parkin zone: mitophagy without Parkin. Trends Cell Biol 28(11):882–895. https://doi.org/10.1016/j.tcb.2018.07.004
Montava-Garriga L, Ganley IG (2019) Outstanding questions in mitophagy: what we do and do not know. J Mol Biol 432(1):206–230. https://doi.org/10.1016/j.jmb.2019.06.032
Youle RJ, Van Der Bliek AM (2012) Mitochondrial fission, fusion, and stress. Science 337(6098):1062–1065. https://doi.org/10.1126/science.1219855
Peng J, Ren KD, Yang J et al (2016) Mitochondrial E3 ubiquitin ligase 1: a key enzyme in regulation of mitochondrial dynamics and functions. Mitochondrion 28:49–53. https://doi.org/10.1016/j.mito.2016.03.007
Strand NS, Allen JM, Ghulam M, Taylor, et al (2018) Dissecting the function of Cullin-RING ubiquitin ligase complex genes in planarian regeneration. Dev Biol 433(2):210–217. https://doi.org/10.1016/j.ydbio.2017.10.011
Higgins GM, Anderson RM (1931) Experimental pathology of the liver –restoration of the liver of the white rat following partial surgical removal. Arch Pathol 12:186–202
Tang TX, Hashimoto T, Chao LY et al (1997) Effects of partial pancreatectomy on liver regeneration in rats. J Surg Res 72(1):8–14. https://doi.org/10.1006/jsre.1997.5165
Uyanoglu M, Canbek M, Aral E et al (2008) Effects of carvacrol upon the liver of rats undergoing partial hepatectomy. Phytomedicine 15(3):226–229. https://doi.org/10.1016/j.phymed.2007.06.010
Gerlach C, Sakkab DY, Scholzen T et al (1997) Ki-67 expression during rat liver regeneration after partial hepatectomy. Hepatology 26(3):573–578. https://doi.org/10.1053/jhep.1997.v26.pm0009303485
Vandesompele J, De Preter K, Pattyn F et al (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3(7):1–12. https://doi.org/10.1186/gb-2002-3-7-research0034
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta ∆C(T)) method. Methods 408:402–408. https://doi.org/10.1006/meth.2001.1262
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3(6):1101–1108. https://doi.org/10.1038/nprot.2008.73
Tanaka T, Arai M, Minemura S et al (2014) Expression level of sonic hedgehog correlated with the speed of gastric mucosa regeneration in artificial gastric ulcers. J Gastroenterol Hepatol 29(4):736–741. https://doi.org/10.1111/jgh.12445
Yu J, Gu X, Yi S (2016) Ingenuity pathway analysis of gene expression profiles in distal nerve stump following nerve injury: insights into Wallerian degeneration. Front Cell Neurosci 10(274):1–12. https://doi.org/10.3389/fncel.2016.00274
Rychtrmoc D, Hubálková L, Víšková A et al (2012) Transcriptome temporal and functional analysis of liver regeneration termination. Physiol Res 61(2):77–92. https://doi.org/10.33549/physiolres.932393
Fujimoto A, Totoki Y, Abe T et al (2012) Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet 44(7):760–764. https://doi.org/10.1038/ng.2291
Motomura M, Kwon KM, Suh SJ et al (2008) Propolis induces cell cycle arrest and apoptosis in human leukemic U937 cells through Bcl-2/Bax regulation. Environ Toxicol Pharmacol 26:61–67. https://doi.org/10.1016/j.etap.2008.01.008
Bruno S, Darzynkiewicz Z (1992) Cell cycle dependent expression and stability of the nuclear protein detected by Ki-67 antibody in HL-60 cells. Cell Proliferat 25(1):31–40. https://doi.org/10.1111/j.1365-2184.1992.tb01435.x
Zhang J, Wan L, Dai X et al (1845) (2014) Functional characterization of anaphase promoting complex/cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. BBA-Rev Cancer 2:277–293. https://doi.org/10.1016/j.bbcan.2014.02.001
Caestecker KW, Van de Walle GR (2013) The role of BRCA1 in DNA double-strand repair: past and present. Exp Cell Res 319(5):575–587. https://doi.org/10.1016/j.yexcr.2012.11.013
Sivakumar S, Gorbsky GJ (2015) Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Bio 16(2):82–94. https://doi.org/10.1038/nrm3934
Ceccaldi R, Sarangi P, D’Andrea AD (2016) The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Bio 17(6):337–349. https://doi.org/10.1038/nrm.2016.48
Macleod KF (2020) Mitophagy and mitochondrial dysfunction in cancer. Annu Rev Cancer Biol 4:41–60. https://doi.org/10.1146/annurev-cancerbio-030419-033405
Di Bacco A, Gill G (2006) SUMO-specific proteases and the cell cycle: an essential role for SENP5 in cell proliferation. Cell Cycle 5(20):2310–2313. https://doi.org/10.4161/cc.5.20.3367
Lee L, Oliva ABP, Churikov D et al (2019) UFMylation of MRE11 is essential for maintenance of telomere length and hematopoietic stem cell survival. Sci Adv. https://doi.org/10.1101/846477
Zhi X, Chen C (2012) WWP1: A versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci 69(9):1425–1434. https://doi.org/10.1007/s00018-011-0871-7
Cheung KF, Lam CNY, Wu K et al (2012) Characterization of the gene structure, functional significance, and clinical application of RNF180, a novel gene in gastric cancer. Cancer 118(4):947–959. https://doi.org/10.1002/cncr.26189
Yang H, Wu L, Ke S et al (2016) Downregulation of ubiquitin-conjugating enzyme UBE2D3 promotes telomere maintenance and radioresistance of Eca-109 human esophageal carcinoma cells. J Cancer 7(9):1152–1162. https://doi.org/10.7150/jca.14745
So CC, Ramachandran S, Martina A (2019) E3 Ubiquitin Ligases RNF20 and RNF40 are required for double-stranded break (DSB) repair: evidence for monoubiquitination of histone H2B lysine 120 as a novel axis of DSB signaling and repair. Mol Cell Biol 39(8):1–19. https://doi.org/10.1128/MCB.00488-18
Liao Y, Shikapwashya ON, Shteyer E et al (2004) Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J Biol Chem 279(41):43107–43116. https://doi.org/10.1074/jbc.M407969200
Yamaji S, Zhang M, Zhang J et al (2010) Hepatocyte-specific deletion of DDB1 induces liver regeneration and tumorigenesis. PNAS 107(51):22237–22242. https://doi.org/10.1073/pnas.1015793108
Cheng R, Liang X, Zhao Q et al (2017) APC/Cdh1 controls cell cycle entry during liver regeneration. Exp Cell Res 354(2):78–84. https://doi.org/10.1016/j.yexcr.2017.03.038
Bachofner M, Speicher T, Bogorad RL et al (2017) Large-scale quantitative proteomics identifies the ubiquitin ligase Nedd4-1 as an essential regulator of liver regeneration. Dev Cell 42:616–625. https://doi.org/10.1016/j.devcel.2017.07.025
Ozeki A, Tsukamoto I (1999) Retinoic acid repressed the expression of c- fos and c-jun and induced apoptosis in regenerating rat liver after partial hepatectomy. BBA-Mol Cell Res 1450:308–319. https://doi.org/10.1016/S0167-4889(99)00063-4
Starkel P, De Saeger C, Sempoux C et al (2005) Blunted DNA synthesis and delayed S-phase entry following inhibition of Cdk2 activity in the regenerating rat liver. Lab Invest 85(4):562–571. https://doi.org/10.1038/labinvest.3700245
Steer CJ (1995) Liver regeneration. FASEB 9:1396–1400. https://doi.org/10.1016/B978-1-4160-3258-8.50007-3
Mizutani T, Yokoyama Y, Kokuryo T et al (2013) Calcitonin gene e related peptide regulates the early phase of liver regeneration. J Surg Res 183(1):138–145. https://doi.org/10.1016/j.jss.2012.11.028
Meier M, Andersen KJ, Knudsen AR et al (2016) Liver regeneration is dependent on the extent of hepatectomy. J Surg Res 6:5–7. https://doi.org/10.1016/j.jss.2016.06.020
Matot I, Nachmansson N, Duev O et al (2018) Impaired liver regeneration after hepatectomy and bleeding is associated with a shift from hepatocyte proliferation to hypertrophy. FASEB J 31(12):5283–5295. https://doi.org/10.1096/fj.201700153R
Zou Y, Bao Q, Kumar S et al (2012) Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS ONE 7(2):e30675. https://doi.org/10.1371/journal.pone.0030675
Lehmann K, Tschuor C, Rickenbacher A et al (2012) Liver failure after extended hepatectomy in mice is mediated by a p21-dependent barrier to liver regeneration. Gastroenterology 143(6):1609-1619.e4. https://doi.org/10.1053/j.gastro.2012.08.043
Pines J (2011) Cubism and the cell cycle: The many faces of the APC/C. Nat Rev Mol Cell Bio 12(7):427–438. https://doi.org/10.1038/nrm3132
de Boer HR, Guerrero Llobet S, van Vugt MA (2016) Controlling the response to DNA damage by the APC/C-Cdh1. Cell Mol Life Sci 73(5):949–960. https://doi.org/10.1007/s00018-015-2096-7
Malumbres M, Barbacid M (2009) Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9(3):153–166. https://doi.org/10.1038/nrc2602
Musacchio A (2015) The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25(20):R1002–R1018. https://doi.org/10.1016/j.cub.2015.08.051
Gmachl M, Gieffers C, Podtelejnikov AV et al (2000) The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. PNAS 97(16):8973–8978. https://doi.org/10.1073/pnas.97.16.8973
Tang Z, Li B, Bharadwaj R et al (2001) APC2 cullin protein and APC11 RING protein comprise the minimal ubiquitin ligase module of the anaphase-promoting complex. Mol Biol Cell 12(12):3839–3851. https://doi.org/10.1091/mbc.12.12.3839
Xie C, Powell C, Yao M et al (2014) Ubiquitin-conjugating enzyme E2C: a potential cancer biomarker. Int J Biochem Cell B 47:113–117. https://doi.org/10.1016/j.biocel.2013.11.023
Okamoto Y, Ozaki T, Miyazaki K et al (2003) UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res 63(14):4167–4173
Ma R, Kang X, Zhang G et al (2016) High expression of UBE2C is associated with the aggressive progression and poor outcome of malignant glioma. Oncol Lett 11(3):2300–2304. https://doi.org/10.3892/ol.2016.4171
Walker A, Acquaviva C, Matsusaka T et al (2008) UbcH10 has a rate-limiting role in G1 phase but might not act in the spindle checkpoint or as part of an autonomous oscillator. J Cell Sci 121(14):2319–2326. https://doi.org/10.1242/jcs.031591
Van Ree JH, Jeganathan KB, Malureanu L et al (2010) Overexpression of the E2 ubiquitin-conjugating enzyme Ubch10 causes chromosome missegregation and tumor formation. J Cell Biol 188(1):83–100. https://doi.org/10.1083/jcb.200906147
Arvand A, Bastians H, Welford SM et al (1998) EWS/FLI1 up regulates mE2-C, a cyclin-selective ubiquitin conjugating enzyme involved in cyclin B destruction. Oncogene 17(16):2039–2045. https://doi.org/10.1038/sj.onc.1202129
Moldovan GL, D’Andrea AD (2009) How the fanconi anemia pathway guards the genome. Annu Rev Genet 43:223–249. https://doi.org/10.1146/annurev-genet-102108-134222
Jones MJK, Huang TT (2012) The Fanconi anemia pathway in replication stress and DNA crosslink repair. Cell Mol Life Sci 69(23):3963–3974. https://doi.org/10.1007/s00018-012-1051-0
Fang S, Lorick KL, Jensen JP et al (2003) RING finger ubiquitin protein ligases: implications for tumorigenesis, metastasis and for molecular targets in cancer. Semin Cancer Biol 13(1):5–14. https://doi.org/10.1016/S1044-579X(02)00095-0
Zeman MK, Cimprich KA (2014) Causes and consequences of replication stress. Nat Cell Biol 16(1):2–9. https://doi.org/10.1038/ncb2897
Wang GP, Xu CS (2011) Alterations in DNA repair gene expression and their possible regulation in rat-liver regeneration. Genet Mol Biol 34(2):304–309. https://doi.org/10.1590/S1415-47572011005000013
He W, Xiao L, Cao C et al (2016) UBE2T promotes nasopharyngeal carcinoma cell proliferation, invasion, and metastasis by activating the AKT/GSKβ/β-catenin pathway. Oncotarget 7(12):15161–15172. https://doi.org/10.18632/oncotarget.7805
Vaughn JP, Davis PL, Jarboe MD et al (1996) BRCA1 expression is induced before DNA synthesis in both normal and tumor-derived breast cells. Cell Growth Differ 7(6):711–715
Lemasters JJ (2005) Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuv Res 8(1):3–5. https://doi.org/10.1089/rej.2005.8.3
Ferri D, Moro L, Mastrodonato M et al (2005) Ultrastructural zonal heterogeneity of hepatocytes and mitochondria within the hepatic acinus during liver regeneration after partial hepatectomy. Biol Cell 97(4):277–288. https://doi.org/10.1042/bc20040154
Guerrieri F, Vendemiale G, Grattagliano I et al (1999) Mitochondrial oxidative alterations following partial hepatectomy. Free Radic Bio Med 26(1–2):34–41. https://doi.org/10.1016/S0891-5849(98)00145-2
Guerrieri F, Muolo L, Cocco T et al (1995) Correlation between rat liver regeneration and mitochondrial energy metabolism. Biochim Biophys Acta 1268:209–213. https://doi.org/10.1016/0925-4439(95)00072-c
Lin CW, Chen YS, Lin CC et al (2015) Amiodarone as an autophagy promoter reduces liver injury and enhances liver regeneration and survival in mice after partial hepatectomy. Sci Rep-UK 5:15807. https://doi.org/10.1038/srep15807
Palikaras K, Lionaki E, Tavernarakis N (2018) Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20(9):1013–1022. https://doi.org/10.1038/s41556-018-0176-2
Fabbro M, Savage K, Hobson K et al (2004) BRCA1-BARD1 complexes are required for P53Ser-15 phosphorylation and a G1/S arrest following ionizing radiation-induced DNA damage. J Biol Chem 279(30):31251–31258. https://doi.org/10.1074/jbc.M405372200
Eifler K, Vertegaal ACO (2015) SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem Sci 40(12):779–793. https://doi.org/10.1016/j.tibs.2015.09.006
Sarangi P, Zhao X (2015) SUMO-mediated regulation of DNA damage repair and responses. Trends Biochem Sci 40(4):233–242. https://doi.org/10.1016/j.tibs.2015.02.006
Qin B, Yu J, Nowsheen S et al (2019) UFL1 promotes histone H4 ufmylation and ATM activation. Nat Commun 10:1242. https://doi.org/10.1038/s41467-019-09175-0
Lam SY, Murphy C, Foley LA et al (2014) The human ubiquitin conjugating enzyme UBE2J2 (Ubc6) is a substrate for proteasomal degradation. Biochem Bioph Res Co 451(3):361–366. https://doi.org/10.1016/j.bbrc.2014.07.099
Oh E, Akopian D, Rape M (2018) Principles of ubiquitin- dependent signaling. Annu Rev Cell Dev Bi 34:137–162. https://doi.org/10.1146/annurev-cellbio-100617-062802
Li Y, Zhou Z, Chen C (2008) WW domain-containing E3 ubiquitin protein ligase 1 targets p63 transcription factor for ubiquitin-mediated proteasomal degradation and regulates apoptosis. Cell Death Differ 15(12):1941–1951. https://doi.org/10.1038/cdd.2008.134
Hu J, Cui F, Xv Z et al (2020) PNO1 promotes cell proliferation in prostate cancer. https://doi.org/10.21203/rs.2.17848/v3
Watson ER, Brown NG, Peters JM et al (2019) Posing the APC/C E3 ubiquitin ligase to orchestrate cell division. Trends Cell Biol 29(2):117–134. https://doi.org/10.1016/j.tcb.2018.09.007
Zhang Y, Zhou H (2020) LncRNA BCAR4 promotes liver cancer progression by upregulating ANAPC11 expression through sponging miR-1261. Int J Mol Med 46(1):159–166. https://doi.org/10.3892/ijmm.2020.4586
Yi Q, Liu Z, Zhang K et al (2021) The role of long non-coding RNA BCAR4 in human cancers. Hum Cell 34:1301–1309. https://doi.org/10.1007/s13577-021-00556-6
Lee YR, Chen M, Lee JD et al (2019) Reactivation of PTEN tumor suppressor for cancer treatment through inhibition of a MYC-WWP1 inhibitory pathway. Science 364:6441-eaau0159. https://doi.org/10.1126/science.aau0159
Komuro A, Imamura T, Saitoh M et al (2004) Negative regulation of transforming growth factor-ß (TGF-ß) signaling by WW domain-containing protein 1 (WWP1). Oncogene 23:6914–6923. https://doi.org/10.1038/sj.onc.1207885
Udali S, Guarini P, Ruzzenente A et al (2015) DNA metylation and gene expression profiles show novel regulatory pathways in hepatocellular carcinoma. Clin Epigenetics 7(43):1–13. https://doi.org/10.1186/s13148-015-0077-1
Zheng X, Chen K, Liu X et al (2018) High RNF40 expression indicates poor prognosis of hepatocellular carcinoma. Int J Clin Exp Pathol 11(5):2901–2906
Guan GG, Wang WB, Lei BX et al (2015) UBE2D3 is a positive prognostic factor and is negatively correlated with hTERT expression in esophageal cancer. Oncol Lett 9(4):1567–1574. https://doi.org/10.3892/ol.2015.2926
Acknowledgements
Thanks to Prof. Dr. Didem TURGUT COŞAN for her valuable contributions
Funding
This study is funded by Eskisehir Osmangazi University Project Office, Project number 201719040.
Author information
Authors and Affiliations
Contributions
AOY Writing the article, Gene expression analysing, experimental design, Western blotting, rats surgery. MC Funding acquisition, project administration, Supervision, doing histological section and participated in discussion.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ozmen Yaylaci, A., Canbek, M. The role of ubiquitin signaling pathway on liver regeneration in rats. Mol Cell Biochem 478, 131–147 (2023). https://doi.org/10.1007/s11010-022-04482-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-022-04482-5