Abstract
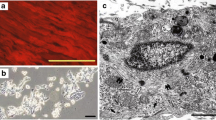
The embryonic rat ventricle H9c2 cells maintain a proliferative state (P condition) in the presence of 10% FCS. However, by reducing serum concentration and in the presence of retinol acetate, proliferation is stopped, myogenic transdifferentiation is inhibited while cardiac differentiation is preserved (D condition). Two-dimensional gel electrophoresis and mass spectrometry analysis was used to define the modifications of the nuclear proteome occurring during the P-to-D transition. Among the proteins observed as modified, a reduced expression of the SM22/transgelin protein was associated with the D state. Also SM22 mRNA levels were reduced during P-to-D transition. Cell transfection experiments indicated that this decrease was partially due to a reduction of the SM22 promoter activity. GATA-4 had a repressive effect on SM22 promoter activity. Thus, since GATA-4 is known as a target of retinoids and may act as a transcriptional repressor, a mechanism to explain the SM22 reduction during the P-to-D transition is tentatively proposed. Immunohistochemical studies on heart cells confirmed the nuclear localization of SM22. Moreover, a differential expression of this protein in different districts of the human heart embryo was detected. Therefore, these data suggest that SM22 expression is regulated during heart development.





Similar content being viewed by others
Abbreviations
- 2-DE:
-
Bi-dimensional electrophoresis
- MALDI-TOF:
-
Matrix assisted laser desorption ionization-time of flight
- MS:
-
Mass spectrometry
- μLC-ESI-IT-MS/MS:
-
Micro-liquid chromatography-electrospray-ion trap-tandem mass spectrometry
- SM22:
-
Transgelin
References
Bruneau BG (2002) Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res 90(5):509–519. doi:10.1161/01.RES.0000013072.51957.B7
Sachinidis A, Fleischmann BK, Kolossov E et al (2003) Cardiac specific differentiation of mouse embryonic stem cells. Cardiovasc Res 58(2):278–291. doi:10.1016/S0008-6363(03)00248-7
Brand T (2003) Heart development: molecular insights into cardiac specification and early morphogenesis. Dev Biol 258(1):1–19. doi:10.1016/S0012-1606(03)00112-X
van der Heyden MA, Defize LH (2003) Twenty one years of P19 cells: what an embryonal carcinoma cell line taught us about cardiomyocyte differentiation. Cardiovasc Res 58(2):292–302. doi:10.1016/S0008-6363(02)00771-X
Puceat M (2005) Role of Rac-GTPase and reactive oxygen species in cardiac differentiation of stem cells. Antioxid Redox Signal 7(11–12):1435–1439. doi:10.1089/ars.2005.7.1435
Brunskill EW, Witte DP, Yutzey KE et al (2001) Novel cell lines promote the discovery of genes involved in early heart development. Dev Biol 235(2):507–520. doi:10.1006/dbio.2001.0313
Kimes BW, Brandt BL (1976) Properties of a clonal muscle cell line from rat heart. Exp Cell Res 98(2):367–381. doi:10.1016/0014-4827(76)90447-X
Merten KE, Jiang Y (2006) Calcineurin activation is not necessary for doxorubicin-induced hypertrophy in H9c2 embryonic rat cardiac cells: involvement of the phosphoinositide 3-kinase-Akt pathway. J Pharmacol Exp Ther 319:934–940. doi:10.1124/jpet.106.108845
Khaw BA, Torchilin VP, Vural I et al (1995) Prevention of hypoxic cardiocyte death by sealing membrane lesions with antimyosin-liposomes. Nat Med 1:1195–1198. doi:10.1038/nm1195-1195
Wu ML, Tsai KL, Wang SM et al (1996) Mechanism of hydrogen peroxide and hydroxyl free radical-induced intracellular acidification in cultured rat cardiac myoblast. Circ Res 78:564–572
Pagano M, Naviglio S (2004) Differentiation of H9c2 cardiomyoblasts: the role of adenylate cyclase system. J Cell Physiol 198:408–416. doi:10.1002/jcp.10420
Hong F, Moon K, Kim SS et al (2001) Role of phospholipase Cgamma1 in insulin-like growth factor I-induced muscle differentiation of H9c2 cardiac myoblasts. Biochem Biophys Res Commun 202:816–822. doi:10.1006/bbrc.2001.4644
Mènard C, Pupier S, Mornet D et al (1999) Modulation of L-type calcium channel expression during retinoic acid-induced differentiation of H9c2 cardiac cells. J Biol Chem 274(41):29063–29070. doi:10.1074/jbc.274.41.29063
Cluzeaut F, Maurer-Schultze B (1986) Proliferation of cardiomyocytes and interstitial cells in the cardiac muscle of the mouse during pre- and postnatal development. Cell Tissue Kinet 19(3):267–274
Kim SH, Kim HS, Lee MM (2002) Re-expression of fetal troponin isoforms in the postinfarction failing heart of the rat. Circ J 66:959–964. doi:10.1253/circj.66.959
Simpson RJ, Dorow DS (2001) Cancer proteomics: from signalling networks to tumor markers. Trends Biotechnol 19(10 (suppl)):40–48. doi:10.1016/S0167-7799(01)01801-7
Li L, Miano JM, Mercer B et al (1996) Expression of the SM22alpha promoter in transgenic mice provides evidence for distinct transcriptional regulatory programs in vascular and visceral smooth muscle cells. J Cell Biol 132(5):849–859. doi:10.1083/jcb.132.5.849
Dentice M, Morisco C, Vitale M (2003) The different cardiac expression of the type 2 iodothyronine deiodinase gene between human and rat is related to the differential response of the Dio2 genes to Nkx-2.5 and GATA-4 transcription factors. Mol Endocrinol 17(8):1508–1521. doi:10.1210/me.2002-0348
Francio-Lang H, Zannini M, De Felice M et al (1992) Multiple mechanisms of interference between transformation and differentiation in thyroid cells. Mol Cell Biol 12(12):5793–5800
Thiede B, Dimmler C, Siejak F et al (2001) Predominant identification of RNA-binding proteins in Fas-induced apoptosis by proteome analysis. J Biol Chem 276(28):26044–26050. doi:10.1074/jbc.M101062200
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72(7):248–254. doi:10.1016/0003-2697(76)90527-3
Vascotto C, Cesarotto L, D’Ambrosio C et al (2006) Proteomic analysis of liver tissues subjected to early ischemia/reperfusion injury during human orthotopic liver transplantation. Proteomics 6(11):3455–3465. doi:10.1002/pmic.200500770
Talamo F, D’Ambrosio C, Arena S et al (2003) Proteins from bovine tissues and biological fluids: defining a reference electrophoresis map for liver, kidney, muscle, plasma and red blood cells. Proteomics 3(4):440–460. doi:10.1002/pmic.200390059
Arena S, D’Ambrosio C, Renzone G et al (2006) A study of Streptococcus thermophilus proteome by integrated analytical procedures and differential expression investigations. Proteomics 6(1):181–192. doi:10.1002/pmic.200402109
Rocco M, D’Ambrosio C, Arena S et al (2006) Proteomic analysis of tomato fruits from two ecotypes during ripening. Proteomics 6(13):3781–3791. doi:10.1002/pmic.200600128
Zhang W, Chait BT (2000) ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal Chem 72(11):2482–2489. doi:10.1021/ac991363o
Qian WJ, Liu T, Monroe ME et al (2005) Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res 4(1):53–62. doi:10.1021/pr0498638
Ralhan R, Desouza LV, Matta A et al (2008) Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol Cell Proteomics 7(6):1162–1173. doi:10.1074/mcp.M700500-MCP200
Rehman I, Azzouzi AR, Cross S et al (2004) Dysregulated expression of S100A11 (calgizzarin) in prostate cancer and precursor lesions. Hum Pathol 35(11):1385–1391. doi:10.1016/j.humpath.2004.07.015
Ikeda R, Yoshida K, Ushiyama M et al (2006) The small heat shock protein alphaB-crystallin inhibits differentiation-induced caspase 3 activation and myogenic differentiation. Biol Pharm Bull 29(9):1815–1819. doi:10.1248/bpb.29.1815
Hoffrogge R, Mikkat S, Scharf C et al (2006) 2-DE proteome analysis of a proliferating and differentiating human neuronal stem cell line (ReNcell VM). Proteomics 6(6):1833–1847. doi:10.1002/pmic.200500556
Shi YY, Wang HC, Yin YH et al (2005) Identification and analysis of tumour-associated antigens in hepatocellular carcinoma. Br J Cancer 92(5):929–934. doi:10.1038/sj.bjc.6602460
Lawson D, Harrison M, Shapland C (1997) Fibroblast transgelin and smooth muscle SM22alpha are the same protein, the expression of which is down-regulated in many cell lines. Cell Motil Cytoskeleton 38(3):250–257. doi:10.1002/(SICI)1097-0169(1997)38:3<250::AID-CM3>3.0.CO;2-9
Assinder SJ, Stanton JA, Prasad PD (2009) Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol 41(3):482–486
Shields JM, Rogers-Graham K, Der CJ (2002) Loss of transgelin in breast and colon tumours and in RIE-1 cells by Ras deregulation of gene expression through Raf-independent pathways. J Biol Chem 277(12):9790–9799. doi:10.1074/jbc.M110086200
Wulfkuhle JD, Sgroi DC, Krutzsch H et al (2002) Proteomics of human breast ductal carcinoma in situ. Cancer Res 62(22):6740–6749
Huang Q, Huang Q, Chen W et al (2008) Identification of transgelin as a potential novel biomarker for gastric adenocarcinoma based on proteomics technology. J Cancer Res Clin Oncol. doi:10.1007/s00432-008-0398-y
Arceci RJ, King AA, Simon MC et al (1993) Mouse GATA-4: a retinoic acid-inducible GATA-binding transcription factor expressed in endodermally derived tissues and heart. Mol Cell Biol 13(4):2235–2246
Molkentin JD, Kalvakolanu DV, Markham BE (1994) Transcription factor GATA-4 regulates cardiac muscle-specific expression of the α-Myosin heavy-chain gene. Mol Cell Biol 14(7):4947–4957
Murakami A, Ishida S, Dickson C (2002) GATA-4 interacts distinctively with negative and positive regulatory elements in the Fgf-3 promoter. Nucleic Acids Res 30(4):1056–1064. doi:10.1093/nar/30.4.1056
Wang L, Tanaka S, Ramirez F (2005) GATA-4 binds to an upstream element of the human alpha2(I)collagen gene (COL1A2) and inhibits transcription in fibroblasts. Matrix Biol 24(5):333–340. doi:10.1016/j.matbio.2005.06.001
Shapland C, Hsuan JJ, Totty NF et al (1993) Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J Cell Biol 121(5):1065–1073. doi:10.1083/jcb.121.5.1065
Moessler H, Mericskay M, Li Z et al (1996) The SM22 promoter directs tissue-specific expression in arterial but not in venous or visceral smooth muscle cells in transgenic mice. Development 122(8):2415–2425
Gerdes J (1990) Ki-67 and other proliferation markers useful for immunohistological diagnostic and prognostic evaluation in human malignancies. Semin Cancer Biol 1(3):199–206
Pan J, Baker KM (2007) Retinoic acid and the heart. Vitam Horm 75:257–283. doi:10.1016/S0083-6729(06)75010-5
Acknowledgments
This work is funded by grants from MIUR to GD (PRIN No. 2007N8P32H_002), GT (FIRB No. RBRN07BMCT—Italian Human ProteomeNet) and AS (MERIT) and from CNR (AG.P04.015 and RSTL 862). Expression vector GATA-4 was provided by Prof. D. Salvatore (University Federico II, Naples).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bregant, E., Renzone, G., Lonigro, R. et al. Down-regulation of SM22/transgelin gene expression during H9c2 cells differentiation. Mol Cell Biochem 327, 145–152 (2009). https://doi.org/10.1007/s11010-009-0052-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-009-0052-2