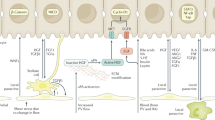
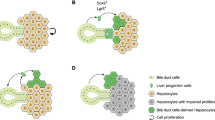
Abstract
Liver possesses the capacity to restore its tissue mass and attain optimal volume in response to physical, infectious and toxic injury. The extraordinary ability of liver to regenerate is the effect of cross-talk between growth factors, cytokines, matrix components and many other factors. In this review we present recent findings and existing information about mechanisms that regulate liver growth, paying attention to augmenter of liver regeneration.
Similar content being viewed by others
References
Fausto N: Liver regeneration. J Hepatol 32: 19–31, 2000
Court FG, Wemyss-Holden SA, Dennison AR, Maddern GJ: The mystery of liver regeneration. Br J Surg 89: 1089–1095, 2002
Nagasue N, Yukaya H, Ogawa Y, Kohno H, Nakamura T: Human liver regeneration after major hepatic resection. A study of normal liver and livers with chronic hepatitis and cirrhosis. Ann Surg 206: 30–39, 1987
Chang TH, Hakamada K, Toyoki Y, Tsuchida S, Sasaki M: Expression of MRP2 and MRP3 during liver regeneration after 90% partial hepatectomy in rats. Transplantation 77: 22–27, 2004
Kurumiya Y, Nozawa K, Sakaguchi K, Nagino M, Nimura Y, Yoshida S: Differential suppression of liver-specific genes in regenerating rat liver induced by extended hepatectomy. J Hepatol 32: 636–644, 2000
Fukuchi T, Hirose H, Onitsuka A, Hayashi M, Senga S, Imai N, Shibata M, Yamauchi K, Futamura N, Sumi Y: Effects of portal-systemic shunt following 90% partial hepatectomy in rats. J Surg Res 89: 126–131, 2000
Steer CJ: Liver regeneration. FASEB J 9: 1396–1400, 1995
Diehl AM, Rai RM: Regulation of signal transduction during liver regeneration. FASEB J 10: 215–227, 1996
LaBrecque D: Liver regeneration: a picture emerges from the puzzle. Am J Gastroenterol 89: 86–96, 1994
Michalopoulos GK, DeFrances MC: Liver regeneration. Science 276: 60–66, 1997
Rozga J: Hepatocyte proliferation in health and in liver failure. Med Sci Monit 8: 32–38, 2002
Michalopoulos GK: Liver regeneration: molecular mechanism of growth control. FASEB J 4: 176–187, 1990
Melchiorri C, Chieco P, Zedda AI, Coni P, Ledda-Columbano GM, Columbano A: Ploidy and nuclearity of rat hepatocytes after compensatory regeneration or mitogen-induced liver growth. Carcinogenesis 14: 1825–1830, 1993
Minuk GY: Hepatic regeneration: if it ain't broke, don't fix it. Can J Gastroenterol 17: 418–424, 2003
Fausto N, Laird AD, Weber EM: Role of growth factors and cytokines in hepatic regeneration. FASEB J 9: 1527–1536, 1995
Zimmermann A: Regulation of liver regeneration. Nephrol Dial Transplant 19: 6–10, 2004
Webber EM, Godowski PJ, Fausto N: In vivo response of hepatocytes to growth factors requires an initial priming stimulus. Hepatology 19: 489–497, 1994
Fausto N, Mead JE, Braun L, Thompson NL, Panzica M, Goyette M, Bell GI, Shank PR: Proto-oncogene expression and growth factors during liver regeneration. Symp Fundam Cancer Res 39: 69–86, 1986
Su AI, Guidotti LG, Pezacki JP, Chisari FV, Schultz PG: Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc Natl Acad Sci U S A 99: 11181–11186, 2002
Jaeschke H: Reactive oxygen and mechanisms of inflammatory liver injury. J Gastroenterol Hepatol 15: 718–724, 2000
Guerrieri F, Vendemiale G, Grattagliano I, Cocco T, Pellecchia G, Altomare E: Mitochondrial oxidative alterations following partial hepatectomy. Free Radic Biol Med 26: 34–41, 1999
Markiewski MM, Mastellos D, Tudoran R, DeAngelis RA, Strey CW, Franchini S, Wetsel RA, Erdei A, Lambris JD: C3a and C3b activation products of the third component of complement (C3) are critical for normal liver recovery after toxic injury. J Immunol 173: 747–754, 2004
Strey CW, Markiewski M, Mastellos D, Tudoran R, Spruce LA, Greenbaum LE, Lambris JD: The proinflammatory mediators C3a and C5a are essential for liver regeneration. J Exp Med 198: 913–923, 2003
Bansal MB, Kovalovich K, Gupta R, Li W, Agarwal A, Radbill B, Alvarez CE, Safadi R, Fiel MI, Friedman SL, Taub RA: Interleukin-6 protects hepatocytes from CCl(4)-mediated necrosis and apoptosis in mice by reducing MMP-2 expression. J Hepatol 42: 548–556, 2005
Zimmers TA, McKillop IH, Pierce RH, Yoo JY, Koniaris LG: Massive liver growth in mice induced by systemic interleukin-6 administration. Hepatology 38: 326–334, 2003
Taub R: Hepatoprotection via the IL-6//Stat3 pathway. J Clin Invest 112: 978–980, 2003
Li W, Liang X, Leu JI, Kovalovich K, Ciliberto G, Taub R: Global changes in interleukin-6-dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology 33: 1377–1386, 2001
Hemmann U, Gerhartz C, Heesel B, Sasse J, Kurapkat G, Grotzinger J, Wollmer A, Zhong Z, Darnell JE Jr, Graeve L, Heinrich PC, Horn F: Differential activation of acute phase response factor//Stat3 and Stat1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. II. Src homology SH2 domains define the specificity of stat factor activation. J Biol Chem 271: 12999–3007, 1996
Li W, Liang X, Kellendonk C, Poli V, Taub R: STAT3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem 277: 28411–28417, 2002
Stahl N, Boulton TG, Farruggella T, Ip NY, Davis S, Witthuhn BA, Quelle FW, Silvennoinen O, Barbieri G, Pellegrini S et al.: Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science 263: 92–95, 1994
Kirillova I, Chaisson M, Fausto N: Tumor necrosis factor induces DNA replication in hepatic cells through nuclear factor kappaB activation. Cell Growth Differ 10: 819–828, 1999
Diehl AM, Yang SQ, Yin M, Lin HZ, Nelson S, Bagby G: Tumor necrosis factor-alpha modulates CCAAT//enhancer binding proteins-DNA binding activities and promotes hepatocyte-specific gene expression during liver regeneration. Hepatology 22: 252–261, 1995
Webber EM, Bruix J, Pierce RH, Fausto N: Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology 28: 1226–1234, 1998
Hayashi H, Nagaki M, Imose M, Osawa Y, Kimura K, Takai S, Imao M, Naiki T, Kato T, Moriwaki H: Normal liver regeneration and liver cell apoptosis after partial hepatectomy in tumor necrosis factor-alpha-deficient mice. Liver Int 25: 162–170, 2005
Gallucci RM, Simeonova PP, Toriumi W, Luster MI: TNF-alpha regulates transforming growth factor-alpha expression in regenerating murine liver and isolated hepatocytes. J Immunol 164: 872–878, 2000
Argast GM, Campbell JS, Brooling JT, Fausto N: Epidermal growth factor receptor transactivation mediates tumor necrosis factor-induced hepatocyte replication. J Biol Chem 279: 34530–34536, 2004
Webber EM, Wu JC, Wang L, Merlino G, Fausto N: Overexpression of transforming growth factor-alpha causes liver enlargement and increased hepatocyte proliferation in transgenic mice. Am J Pathol 145: 398–408, 1994
Russell WE, Dempsey PJ, Sitaric S, Peck AJ, Coffey RJ Jr: Transforming growth factor-alpha (TGF alpha) concentrations increase in regenerating rat liver:evidence for a delayed accumulation of mature TGF alpha. Endocrinology 133: 1731–1738, 1993
Skarpen E, Oksvold MP, Grosvik H, Widnes C, Huitfeldt HS: Altered regulation of EGF receptor signaling following a partial hepatectomy. J Cell Physiol 202: 707–716, 2005
Tomiya T, Ogata I, Yamaoka M, Yanase M, Inoue Y, Fujiwara K: The mitogenic activity of hepatocyte growth factor on rat hepatocytes is dependent upon endogenous transforming growth factor-alpha. Am J Pathol 157: 1693–1701, 2000
Tomiya T, Ogata I, Fujiwara K: Transforming growth factor alpha levels in liver and blood correlate better than hepatocyte growth factor with hepatocyte proliferation during liver regeneration. Am J Pathol 153:955–961, 1998
Schmidt C, Bladt, F, Goedecke S, Brinkmann V, Zschiesche W, Sharpe M, Gherardi E, Birchmeier C: Scatter factor/hepatocyte growth factor is essential for liver development. Nature 373: 699–702, 1995
Noji S, Tashiro K, Koyama E, Nohno T, Ohyama K, Taniguchi S, Nakamura T: Expression of hepatocyte growth factor gene in endothelial and Kupffer cells of damaged rat livers, as revealed by in situ hybridization. Biochem Biophys Res Commun 173: 42–47, 1990
Fabregat I, de Juan C, Nakamura T, Benito M: Growth stimulation of rat fetal hepatocytes in response to hepatocyte growth factor: modulation of c-myc and c-fos expression. Biochem Biophys Res Commun 189: 684–690, 1992
Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC: Cross-talk between epidermal growth factor receptor and c-Met signal pathways in transformed cells. J Biol Chem 275: 8806–8811, 2000
Presnell SC, Stolz DB, Mars WM, Jo M, Michalopoulos GK, Strom SC: Modifications of the hepatocyte growth factor/c-met pathway by constitutive expression of transforming growth factor-alpha in rat liver epithelial cells. Mol Carcinog 18: 244–255, 1997
Stolz DB, Michalopoulos GK: Comparative effects of hepatocyte growth factor and epidermal growth factor on motility, morphology, mitogenesis, and signal transduction of primary rat hepatocytes. J Cell Biochem 55: 445–464, 1994
de Juan C, Benito M, Alvarez A, Fabregat I: Differential proliferative response of cultured fetal and regenerating hepatocytes to growth factors and hormones. Exp Cell Res 202: 495–500, 1992
Klein D, Schubert T, Horch RE, Jauch KW, Jeschke MG: Insulin treatment improves hepatic morphology and function through modulation of hepatic signals after severe trauma. Ann Surg 240: 340–349, 2004
Knopp J, Macho L, Fickova M, Zorad S, Kvetnansky R, Jaroscakova I: Insulin and catecholamines act at different stages of rat liver regeneration. Ann N Y Acad Sci 827: 489–493, 1997
Houck KA, Michalopoulos GK: Altered responses of regenerating hepatocytes to norepinephrine and transforming growth factor type beta. J Cell Physiol 141: 503–509, 1989
Francavilla A, Gavaler JS, Makowka L, Barone M, Mazzaferro V, Ambrosino G, Iwatsuki S, Guglielmi FW, Dileo A, Balestrazzi A et al: Estradiol and testosterone levels in patients undergoing partial hepatectomy. A possible signal for hepatic regeneration? Dig Dis Sci 34: 818–822, 1989
Chiu EJ, Lin HL, Chi CW, Liu TY, Lui WY: Estrogen therapy for hepatectomy patients with poor liver function? Med Hypotheses 58: 516–518, 2002
Francavilla A, Eagon PK, DiLeo A, Polimeno L, Panella C, Aquilino AM, Ingrosso M, Van Thiel DH, Starzl TE: Sex hormone-related functions in regenerating male rat liver. Gastroenterology 91: 1263–1270, 1986
Metcalfe AM, Phillips P, Dixon RM, Radda GK: Vasopressin synergistically stimulates DNA synthesis in normal and regenerating rat liver cell cultures in the presence of hepatocyte growth factor. J Mol Endocrinol 18: 161–166, 1997
Pennisi PA, Kopchick JJ, Thorgeirsson S, LeRoith D, Yakar S: Role of growth hormone (GH) in liver regeneration. Endocrinology 145: 4748–4755, 2004
LaBrecque DR, Pesch LA: Preparation and partial characterization of hepatic regenerative stimulator substance (SS) from rat liver. J Physiol 248: 273–284, 1975
Gandhi CR, Kuddus R, Subbotin VM, Prelich J, Murase N, Rao AS, Nalesnik MA, Watkins SC, DeLeo A, Trucco M, Starzl TE: A fresh look at augmenter of liver regeneration in rats. Hepatology 29: 1435–1445, 1999
Wang G, Yang X, Zhang Y, Wang Q, Chen H, Wei H, Xing G, Xie L, Hu Z, Zhang C, Fang D, Wu C, He F: Identification and characterization of receptor for mammalian hepatopoietin that is homologous to yeast ERV1. J Biol Chem 274: 11469–11472, 1999
Liu Q, Yu HF, Sun H, Ma HF: Expression of human augmenter of liver regeneration in pichia pastoris yeast and its bioactivity in vitro. World J Gastroenterol 10: 3188–3190, 2004
Hagiya M, Francavilla A, Polimeno L, Ihara I, Sakai H, Seki T, Shimonishi M, Porter KA, Starzl TE: Cloning and sequence analysis of the rat augmenter of liver regeneration (ALR) gene: expression of biologically active recombinant ALR and demonstration of tissue distribution. Proc Natl Acad Sci U S A 91: 8142–8146, 1994
Cheng J, Zhong YW, Liu Y, Dong J, Yang JZ, Chen JM: Cloning and sequence analysis of human genomic DNA of augmenter of liver regeneration. World J Gastroenterol 6: 275–277, 2000
Giorda R, Hagiya M, Seki T, Shimonishi M, Sakai H, Michaelson J, Francavilla A, Starzl TE, Trucco M: Analysis of the structure and expression of the augmenter of liver regeneration (ALR) gene. Mol Med 2: 97–108, 1996
Wu CK, Dailey TA, Dailey HA, Wang BC, Rose JP: The crystal structure of augmenter of liver regeneration: a mammalian FAD-dependent sulfhydryl oxidase. Protein Sci 12: 1109–1118, 2003
Li Y, Wei K, Lu C, Li Y, Li M, Xing G, Wei H, Wang Q, Chen J, Wu C, Chen H, Yang S, He F: Identification of hepatopoietin dimerization, its interacting regions and alternative splicing of its transcription. Eur J Biochem 269: 3888–3893, 2002
Lu J, Xu WX, Zhan YQ, Cui XL, Cai WM, He FC, Yang XM: Identification and characterization of a novel isoform of hepatopoietin. World J Gastroenterol 8: 353–356, 2002
Lisowsky T, Lee JE, Polimeno L, Francavilla A, Hofhaus G: Mammalian augmenter of liver regeneration protein is a sulfhydryl oxidase. Dig Liver Dis 33: 173–180, 2001
Lee J, Hofhaus G, Lisowsky T: Erv1p from Saccharomyces cerevisiae is a FAD-linked sulfhydryl oxidase. FEBS Lett 477: 62–66, 2000
Levitan A, Danon A, Lisowsky T: Unique features of plant mitochondrial sulfhydryl oxidase. J Biol Chem 279: 20002–20008, 2004
Senkevich TG, White CL, Koonin EV, Moss B: A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc Natl Acad Sci U S A 97: 12068–12073, 2000
Lutz T, Neupert W, Herrmann JM: Import of small Tim proteins into the mitochondrial intermembrane space. EMBO J 22: 4400–4408, 2003
Chacinska A, Pfannschmidt S, Wiedemann N, Kozjak V, Sanjuan Szklarz LK, Schulze-Specking A, Truscott KN, Guiard B, Meisinger C, Pfanner N: Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins. EMBO J 23: 3735–3746, 2004
Rissler M, Wiedemann N, Pfannschmidt S, Gabriel K, Guiard B, Pfanner N, Chacinska A: The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins. J Mol Biol 353: 485–492, 2005
Mesecke N, Terziyska N, Kozany C, Baumann F, Neupert W, Hell K, Herrmann JM: A disulfide relay system in the intermembrane space of mitochondria that mediates protein import. Cell 121: 1059–1069, 2005
Farrell SR, Thorpe C: Augmenter of liver regeneration: a flavin-dependent sulfhydryl oxidase with cytochrome c reductase activity. Biochemistry 44: 1532–1541, 2005
Allen S, Balabanidou V, Sideris DP, Lisowsky T, Tokatlidis K: Erv1 mediates the Mia40-dependent protein import pathway and provides a functional link to the respiratory chain by shuttling electrons to cytochrome c. J Mol Biol 353: 937–944, 2005
Kadokura H, Beckwith J: Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J 21: 2354–2363, 2002
Kadokura H, Bader M, Tian H, Bardwell JC, Beckwith J: Roles of a conserved arginine residue of DsbB in linking protein disulfide-bond-formation pathway to the respiratory chain of Escherichia coli. Proc Natl Acad Sci U S A 97: 10884–10889, 2000
Stein G, Lisowsky T: Functional comparison of the yeast scERV1 and scERV2 genes. Yeast 14: 171–180, 1998
Gerber J, Muhlenhoff U, Hofhaus G, Lill R, Lisowsky T: Yeast ERV2p is the first microsomal FAD-linked sulfhydryl oxidase of the Erv1p/Alrp protein family. J Biol Chem 276(26): 23486–23491, 2001
Gross E, Sevier CS, Vala A, Kaiser CA, Fass D: A new FAD-binding fold and intersubunit disulfide shuttle in the thiol oxidase Erv2p. Nat Struct Biol 9: 61–67, 2002
Sevier CS, Cuozzo JW, Vala A, Aslund F, Kaiser CA: A flavoprotein oxidase defines a new endoplasmic reticulum pathway for biosynthetic disulphide bond formation. Nat Cell Biol 3: 874–882, 2001
Frand AR, Kaiser CA: Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell 4: 469–477, 1999
Tu BP, Weissman JS: Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol 164: 341–346, 2004
Thorpe C, Hoober KL, Raje S, Glynn NM, Burnside J, Turi GK, Coppock DL: Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys 405: 1–12, 2002
Lill R, Muhlenhoff U: Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci 30: 133–141, 2005
Lill R, Kispal G: Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci 25: 352–356, 2000
Lange H, Lisowsky T, Gerber J, Muhlenhoff U, Kispal G, Lill R: An essential function of the mitochondrial sulfhydryl oxidase Erv1p/ALR in the maturation of cytosolic Fe/S proteins. EMBO Rep 2: 715–720, 2001
Klissenbauer M, Winters S, Heinlein UA, Lisowsky T: Accumulation of the mitochondrial form of the sulphydryl oxidase Erv1p/Alrp during the early stages of spermatogenesis. J Exp Biol 205: 1979–1986, 2002
Cheng J, Wang L, Li K, Lu YY, Wang G, Liu Y, Zhong YW, Duan HJ, Hong Y, Li L, Zhang LX, Chen JM: Screening of augmenter of liver regeneration-binding proteins by yeast-two hybrid technique. Hepatobiliary Pancreat Dis Int 2: 81–84, 2003
Tury A, Mariet-Coello G, Lisowsky T, Bernardette G, Fellman D: Expression of the sulfhydryl oxidase ALR (augmenter of liver regeneration) in adult rat brain. Brain Res 1048: 87–97, 2005
Polimeno L, Capuano F, Marangi LC, Margiotta M, Lisowsky T, Ierardi E, Francavilla R, Francavilla A: The augmenter of liver regeneration induces mitochondrial gene expression in rat liver and enhances oxidative phosphorylation capacity of liver mitochondria. Dig Liver Dis 32: 510–517, 2000
Shadel GS, Clayton DA: Mitochondrial transcription initiation. Variation and conservation. J Biol Chem 268: 16083–16086, 1993
Hofhaus G, Stein G, Polimeno L, Francavilla A, Lisowsky T: Highly divergent amino termini of the homologous human ALR and yeast scERV1 gene products define species specific differences in cellular localization. Eur J Cell Biol 78: 349–356, 1999
McGowan JA, Strain AJ, Bucher NL: DNA synthesis in primary cultures of adult rat hepatocytes in a defined medium: effects of epidermal growth factor, insulin, glucagon, and cyclic-AMP. J Cell Physiol. 108: 353–363, 1981
Skov Olsen P, Boesby S, Kirkegaard P, Therkelsen K, Almdal T, Poulsen SS, Nexo E: Influence of epidermal growth factor on liver regeneration after partial hepatectomy in rats. Hepatology 8: 992–996, 1988
Vujanovic NL, Polimeno L, Azzarone A, Francavilla A, Chambers WH, Starzl TE, Herberman RB, Whiteside TL: Changes of liver-resident NK cells during liver regeneration in rats. J Immunol 154: 6324–6338, 1995
Francavilla A, Vujanovic NL, Polimeno L, Azzarone A, Iacobellis A, Deleo A, Hagiya M, Whiteside TL, Starzl TE: The in vivo effect of hepatotrophic factors augmenter of liver regeneration, hepatocyte growth factor, and insulin-like growth factor-II on liver natural killer cell functions. Hepatology 25: 411–415, 1997
Tanigawa K, Sakaida I, Masuhara M, Hagiya M, Okita K: Augmenter of liver regeneration (ALR) may promote liver regeneration by reducing natural killer (NK) cell activity in human liver diseases. J Gastroenterol 35: 112–119, 2000
Polimeno L, Margiotta M, Marangi L, Lisowsky T, Azzarone A, Ierardi E, Frassanito MA, Francavilla R, Francavilla A: Molecular mechanisms of augmenter of liver regeneration as immunoregulator: its effect on interferon-gamma expression in rat liver. Dig Liver Dis 32: 217–225, 2000
Li Y, Xing G, Wang Q, Li M, Wei H, Fan G, Chen J, Yang X, Wu C, Chen H, He F: Hepatopoietin acts as an autocrine growth factor in hepatoma cells. DNA Cell Biol 20: 791–795, 2001
Li Y, Li M, Xing G, Hu Z, Wang Q, Dong C, Wei H, Fan G, Chen J, Yang X, Zhao S, Chen H, Guan K, Wu C, Zhang C, He F: Stimulation of the mitogen-activated protein kinase cascade and tyrosine phosphorylation of the epidermal growth factor receptor by hepatopoietin. J Biol Chem 275: 37443–37447, 2000
An W, Liu XJ, Lei TG, Dai J, Du GG: Growth induction of hepatic stimulator substance in hepatocytes through its regulation on EGF receptors. Cell Res 9: 37–49, 1999
Lu C, Li Y, Zhao Y, Xing G, Tang F, Wang Q, Sun Y, Wei H, Yang X, Wu C, Chen J, Guan KL, Zhang C, Chen H, He F: Intracrine hepatopoietin potentiates AP-1 activity through JAB1 independent of MAPK pathway. FASEB J 16: 90–92, 2002
Claret FX, Hibi M, Dhut S, Toda T, Karin M: A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 383: 453–457, 1996
Naumann M, Bech-Otschir D, Huang X, Ferrell K, Dubiel W: COP9 signalosome-directed c-Jun activation/stabilization is independent of JNK. J Biol Chem 274: 35297–35300, 1999
Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W: A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12: 469–478, 1998
Wang Y, Lu C, Wei H, Wang N, Chen X, Zhang L, Zhai Y, Zhu Y, Lu Y, He F: Hepatopoietin interacts directly with COP9 signalosome and regulates AP-1 activity. FEBS Lett 572: 85–91, 2004
Chen X, Li Y, Wei K, Li L, Liu W, Zhu Y, Qiu Z, He F: The potentiation role of hepatopoietin on activator protein-1 is dependent on its sulfhydryl oxidase activity. J Biol Chem 278: 49022–49030, 2003
Kleemann R, Kapurniotu A, Frank RW, Gessner A, Mischke R, Flieger O, Juttner S, Brunner H, Bernhagen J: Disulfide analysis reveals a role for macrophage migration inhibitory factor (MIF) as thiol-protein oxidoreductase. J Mol Biol 280: 85–102, 1998
Li,Y, Lu C, Xing G, Zhu Y, He F: Macrophage migration inhibitory factor directly interacts with hepatopoietin and regulates the proliferation of hepatoma cell. Exp Cell Res 300: 379–387, 2004
Li,Y, Liu W, Xing G, Tian C, Zhu Y, He F: Direct association of hepatopoietin with thioredoxin constitutes a redox signal transduction in activation of AP-1/NF-kappaB. Cell Signal 17: 985–996, 2005
Thasler WE, Dayoub R, Muehlbauer M, Hellerbrand C, Singer T, Graebe A, Jauch KW, Schlitt HJ, Weiss TS: Repression of cytochrome P450 activity in human hepatocytes in vitro by a novel hepatotrophic factor Augmenter of Liver Regeneration. J Pharmacol Exp Ther (in press)
Thasler WE, Schlott T, Thelen P, Hellerbrand C, Bataille F, Lichtenauer M, Schlitt HJ, Jauch KW, Weiss TS: Expression of augmenter of liver regeneration (ALR) in human liver cirrhosis and carcinoma. Histopathology 47: 57–66, 2005
Li Q, Liu DW, Zhang LM, Zhu B, He YT, Xiao YH: Effects of augmentation of liver regeneration recombinant plasmid on rat hepatic fibrosis. World J Gastroenterol 11: 2438–2443, 2005
LaBrecque DR: Hepatic stimulator substance. Discovery, characteristics and mechanism of action. Dig Dis Sci 36: 669–673, 1991
Francavilla A, Ove P, Polimeno L, Coetzee M, Makowka L, Rose J, Van Thiel DH, Starzl TE: Extraction and partial purification of a hepatic stimulatory substance in rats, mice, and dogs. Cancer Res 47: 5600–5605, 1987
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pawlowski, R., Jura, J. ALR and Liver Regeneration. Mol Cell Biochem 288, 159–169 (2006). https://doi.org/10.1007/s11010-006-9133-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-006-9133-7