Abstract
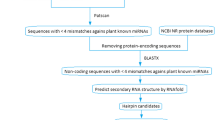
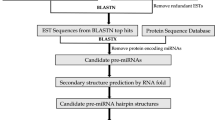
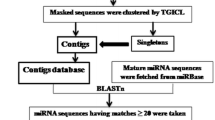
MicroRNAs (miRNAs) are a class of naturally occurring and small non-coding RNA molecules of about 21–25 nucleotides in length. Their main function is to downregulate gene expression in different manners like translational repression, mRNA cleavage and epigenetic modification. To predict new miRNAs in plants different computational approaches have been developed. In the present study, an EST based approach has been used to identify novel miRNAs in horsegram. Identification of miRNAs was initiated by mining the EST database available at NCBI. Total of 989 ESTs were obtained for the identification of miRNAs. These ESTs were subjected to CAP3 assembly to remove the redundancy. This resulted in an output of 72 contigs and 606 singletons as non redundant datasets. The miRNAs were then predicted by using miRNA-finder. A total of eight potential miRNAs were predicted and named as hor-miR1 to hor-miR8. None of identified miRNAs showed significant homology with the previously reported in plants and therefore should be considered novel. These miRNAs were inputted to miRU2 program to predict their targets. The target mRNAs for these miRNAs mainly belong to zinc finger, chromosome condensation, protein kinase, abscisic acid-responsive, calcineurin-like phosphoesterase, disease resistance and transcriptional factor family proteins. These targets appeared to be involved in plant growth and development and environmental stress responses.


Similar content being viewed by others
References
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Nature 391:806–811
Bartel DP (2004) Cell 116:281–297
He L, Hannon GJ (2004) Nat Rev Genet 5:522–531
Brodersen P, Voinnet O (2006) Trend Genet 22:268–280
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) Genes Dev 16:1616–1626
Zuker M (2003) Nucleic Acid Res 31:3406–3415
Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Bioinformatics 20:2911–2917
Chapman EJ, Carrington JC (2007) Nat Rev Genet 8:884–896
Sanan-Mishra N, Mukherjee SK (2007) Open Plant Sci J 1:1–9
Jin H (2008) FEBS Lett 582:2679–2684
Zhu JK (2008) Proc Natl Acad Sci USA 105:9851–9852
Yin Z, Li C, Han X, Shen F (2008) Gene 414:60–66
Sunkar R, Chinnusamy V, Zhu J, Zhu JK (2007) Trends Plant Sci 12:301–309
Mallory AC, Vaucheret H (2006) Nat Genet 38:S31–S36
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) PLoS One 2:e219
Chiou TJ, Aung K, Lin SI, Wu CC, Chiang SF, Su CL (2006) Plant Cell 18:412–421
Jones-Rhoades MW, Bartel DP, Bartel B (2006) Annu Rev Plant Biol 57:19–53
Sunkar R, Kapoor A, Zhu JK (2006) Plant Cell 18:2051–2065
Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK (2005) Curr Biol 15:2038–2043
Lu S, Sun YH, Shi R, Clark C, Li L, Chiang WL (2005) Plant Cell 17:2186–2203
Jones-Rhoades MW, Bartel DP (2004) Mol Cell 14:787–799
Jeswani LM, Baldev B (1990) Advances in pulse production technology publication and information division. Indian Council of Agricultural Research, New Delhi
Yadava ND, Vyas NL (1994) Arid legumes. Agro publishers, India
Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Gen Biol 5:R65
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Science 297:2053–2056
Zhang Y (2005) Nucleic Acid Res 33:W701–W704
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003) RNA 9:277–279
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Plant J 46:243–259
Zhang B, Pan X, Cobb GP, Anderson TA (2006) Dev Biol 289:3–16
Zhao B, Liang R, Ge L, Li W, Xiao H, Lin H, Ruan K, Jin Y (2007) Biochem Biophys Res Commun 354:585–590
Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) Curr Biol 13:807–818
Zhang B, Pan X, Anderson TA (2006) FEBS Lett 580:3753–3762
Zheng Y, Hsu W, Lee M-Li, Wong L (2006) VDMB 4316:131–145
Shi Y, Berg JM (1996) Biochemistry 35:3845–3848
Stone SL, Hauksdottir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Plant Physiol 137:13–30
Ciechanover A (1998) EMBO J 17:7151–7160
Francis NJ, Kingston RE, Woodcock CL (2004) Science 306:1574–1577
Lorkovic ZJ, Barta A (2002) Nucleic Acids Res 30:623–635
Romanel EA, Schrago CG, Counago RM, Russo CA, Alves-Ferreira M (2009) PLoS One 4:e5791
Stefano G, Renna L, Chatre L, Hanton SL, Moreau P, Hawes C, Brandizzi F (2006) Plant J 46:95–110
Deyholos MK, Cavaness GF, Hall B, King E, Punwani J, Van Norman J, Sieburth LE (2003) Development 130:6577–6588
Zhong R, Ye ZH (2004) Plant Cell Physiol 45:1720–1728
Theissen G, Kim JT, Saedler H (1996) J Mol Evol 43:484–516
Boggs NA, Nasrallah JB, Nasrallah ME (2009) PLoS Genet 5:e1000426
Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J (2000) EMBO J 19:542–549
Kobe B, Deisenhofer J (2002) Nature 374:183–1866
Ellis J, Lawrence G, Ayliffe M, Anderson P, Collins N, Finnegan J, Frost D, Luck J, Pryor T (1997) Annu Rev Phytopathol 35:271–291
Johnson KL, Jones BJ, Bacic A, Schultz CJ (2003) Plant Physiol 133:1911–1925
Kalde M, Nuhse TS, Findlay K, Peck SC (2007) Proc Natl Acad Sci USA 104:11850–11855
Shah J (2005) Annu Rev Phytopathol 43:229–260
Acknowledgments
Authors are thankful to Dr. P. S. Ahuja, Director, IHBT for his valuable suggestions and guidance to conduct this work. We would like to thank the financial support from Council of Scientific and Industrial Research (CSIR) and Department of Science and Technology (DST), Govt of India. HM is thankful to CSIR for providing research fellowship in the form of JRF.
Author information
Authors and Affiliations
Corresponding author
Additional information
Jyoti Bhardwaj and Hasan Mohammad contributed equally to this manuscript.
Rights and permissions
About this article
Cite this article
Bhardwaj, J., Mohammad, H. & Yadav, S.K. Computational identification of microRNAs and their targets from the expressed sequence tags of horsegram (Macrotyloma uniflorum (Lam.) Verdc.). J Struct Funct Genomics 11, 233–240 (2010). https://doi.org/10.1007/s10969-010-9098-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10969-010-9098-3