Abstract
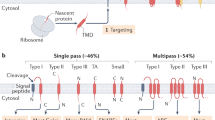
Most membrane proteins are composed of hydrophobic α-helical transmembrane segments and are integrated into the lipid bilayer of the endoplasmic reticulum by the highly conserved Sec61 translocon. With respect to the integration mechanism, three types of transmembrane segments can be distinguished—the signal, the stop-transfer sequence, and the re-integration sequence—which in linear succession can account for all kinds of membrane protein topologies. The transmembrane orientation of the initial signal and to a weaker extent also of downstream transmembrane segments is affected by charged flanking residues according to the so-called positive-inside rule. The main driving force for transmembrane integration is hydrophobicity. Systematic analysis suggested thermodynamic equilibration of each peptide segment in the translocon with the membrane as the underlying mechanism. However, there is evidence that integration is not entirely sequence-autonomous, but depends also on the sequence context, from very closely spaced transmembrane segments to the folding state and properties of neighboring sequences. Topogenesis is even influenced by accessory proteins that appear to act as intramembrane chaperones.





Similar content being viewed by others
Abbreviations
- ER:
-
Endoplasmic reticulum
- Ncyt/Cexo :
-
Cytoplasmic N- and exoplasmic C-terminus
- Nexo/Ccyt :
-
Exoplasmic N- and cytoplasmic C-terminus
- TM:
-
Transmembrane
- TRAM:
-
Translocating chain-associated membrane protein
References
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132. https://doi.org/10.1016/0022-2836(82)90515-0
Wimley WC (2002) Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci 11:301–312. https://doi.org/10.1110/ps.29402
Schiffrin B, Brockwell DJ, Radford SE (2017) Outer membrane protein folding from an energy landscape perspective. BMC Biol 15:123. https://doi.org/10.1186/s12915-017-0464-5
Gruss F, Zähringer F, Jakob RP et al (2013) The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol 20:1318–1320. https://doi.org/10.1038/nsmb.2689
Höhr AIC, Lindau C, Wirth C et al (2018) Membrane protein insertion through a mitochondrial β-barrel gate. Science 359:eaah6834. https://doi.org/10.1126/science.aah6834
Park E, Ménétret J-F, Gumbart JC et al (2014) Structure of the SecY channel during initiation of protein translocation. Nature 506:102–106. https://doi.org/10.1038/nature12720
Gogala M, Becker T, Beatrix B et al (2014) Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 506:107–110. https://doi.org/10.1038/nature12950
Voorhees RM, Hegde RS (2016) Structure of the Sec61 channel opened by a signal sequence. Science 351:88–91. https://doi.org/10.1126/science.aad4992
MacKinnon AL, Paavilainen VO, Sharma A et al (2014) An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. Elife 3:e01483. https://doi.org/10.7554/eLife.01483
Junne T, Wong J, Studer C et al (2015) Decatransin, a novel natural product inhibiting protein translocation at the Sec61/SecY translocon. J Cell Sci 128:1217–1229. https://doi.org/10.1242/jcs.165746
Paatero AO, Kellosalo J, Dunyak BM et al (2016) Apratoxin kills cells by direct blockade of the Sec61 protein translocation channel. Cell Chem Biol 23:561–566. https://doi.org/10.1016/j.chembiol.2016.04.008
Blobel G, Dobberstein B (1975) Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol 67:835–851. https://doi.org/10.1083/jcb.67.3.835
Blobel G (1980) Intracellular protein topogenesis. Proc Natl Acad Sci USA 77:1496–1500
von Heijne G (1990) The signal peptide. J Membr Biol 115:195–201
Heijne G (1986) The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J 5:3021–3027
Nilsson J, Persson B, von Heijne G (2005) Comparative analysis of amino acid distributions in integral membrane proteins from 107 genomes. Proteins 60:606–616. https://doi.org/10.1002/prot.20583
Hartmann E, Rapoport TA, Lodish HF (1989) Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA 86:5786–5790
Szczesna-Skorupa E, Kemper B (1989) NH2-terminal substitutions of basic amino acids induce translocation across the microsomal membrane and glycosylation of rabbit cytochrome P450IIC2. J Cell Biol 108:1237–1243
Beltzer JP, Fiedler K, Fuhrer C et al (1991) Charged residues are major determinants of the transmembrane orientation of a signal-anchor sequence. J Biol Chem 266:973–978
Parks GD, Lamb RA (1991) Topology of eukaryotic type II membrane proteins: importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell 64:777–787. https://doi.org/10.1016/0092-8674(91)90507-U
Denzer AJ, Nabholz CE, Spiess M (1995) Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J 14:6311–6317
Sakaguchi M, Tomiyoshi R, Kuroiwa T et al (1992) Functions of signal and signal-anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc Natl Acad Sci USA 89:16–19. https://doi.org/10.1073/pnas.89.1.16
Wahlberg JM, Spiess M (1997) Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. 137:555–562
Rösch K, Naeher D, Laird V et al (2000) The topogenic contribution of uncharged amino acids on signal sequence orientation in the endoplasmic reticulum. J Biol Chem 275:14916–14922. https://doi.org/10.1074/jbc.M000456200
Goder V, Spiess M (2003) Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J 22:3645–3653. https://doi.org/10.1093/emboj/cdg361
Devaraneni PK, Conti B, Matsumura Y et al (2011) Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell 146:134–147. https://doi.org/10.1016/j.cell.2011.06.004
Kocik L, Junne T, Spiess M (2012) Orientation of internal signal-anchor sequences at the Sec61 translocon. J Mol Biol 424:368–378. https://doi.org/10.1016/j.jmb.2012.10.010
McKenna M, Simmonds RE, High S (2017) Mycolactone reveals the substrate-driven complexity of Sec61-dependent transmembrane protein biogenesis. J Cell Sci 130:1307–1320. https://doi.org/10.1242/jcs.198655
Goder V, Junne T, Spiess M (2004) Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol Biol Cell 15:1470–1478. https://doi.org/10.1091/mbc.E03-08-0599
Junne T, Schwede T, Goder V, Spiess M (2007) Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J Biol Chem 282:33201–33209. https://doi.org/10.1074/jbc.M707219200
Emr SD, Hanley-Way S, Silhavy TJ (1981) Suppressor mutations that restore export of a protein with a defective signal sequence. Cell 23:79–88
Tam PCK, Maillard AP, Chan KKY, Duong F (2005) Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J 24:3380–3388. https://doi.org/10.1038/sj.emboj.7600804
Saparov SM, Erlandson K, Cannon K et al (2007) Determining the conductance of the SecY protein translocation channel for small molecules. Mol Cell 26:501–509. https://doi.org/10.1016/j.molcel.2007.03.022
Flower AM (2007) The SecY translocation complex: convergence of genetics and structure. Trends Microbiol. https://doi.org/10.1016/j.tim.2007.03.001
Audigier Y, Friedlander M, Blobel G (1987) Multiple topogenic sequences in bovine opsin. Proc Natl Acad Sci USA 84:5783–5787. https://doi.org/10.1073/pnas.84.16.5783
Wessels HP, Spiess M (1988) Insertion of a multispanning membrane protein occurs sequentially and requires only one signal sequence. Cell 55:61–70
Lipp J, Flint N, Haeuptle MT, Dobberstein B (1989) Structural requirements for membrane assembly of proteins spanning the membrane several times. J Cell Biol 109:2013–2022. https://doi.org/10.1083/jcb.109.5.2013
Sato M, Hresko R, Mueckler M (1998) Testing the charge difference hypothesis for the assembly of a eucaryotic multispanning membrane protein. J Biol Chem 273:25203–25208. https://doi.org/10.1074/jbc.273.39.25203
Sato M, Mueckler M (1999) A conserved amino acid motif (R-X-G-R-R) in the Glut1 glucose transporter is an important determinant of membrane topology. J Biol Chem 274:24721–24725. https://doi.org/10.1074/jbc.274.35.24721
Gafvelin G, von Heijne G (1994) Topological “frustration” in multispanning E. coli inner membrane proteins. Cell 77:401–412. https://doi.org/10.1016/0092-8674(94)90155-4
Goder V, Bieri C, Spiess M (1999) Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol 147:257–266
Tu L, Khanna P, Deutsch C (2014) Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J Mol Biol 426:185–198. https://doi.org/10.1016/j.jmb.2013.09.013
Hessa T, Kim H, Bihlmaier K et al (2005) Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433:377–381. https://doi.org/10.1038/nature03216
Hessa T, Meindl-Beinker NM, Bernsel A et al (2007) Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 450:1026–1030. https://doi.org/10.1038/nature06387
Hedin LE, Ojemalm K, Bernsel A et al (2010) Membrane insertion of marginally hydrophobic transmembrane helices depends on sequence context. J Mol Biol 396:221–229. https://doi.org/10.1016/j.jmb.2009.11.036
MacCallum JL, Tieleman DP (2011) Hydrophobicity scales: a thermodynamic looking glass into lipid-protein interactions. Trends Biochem Sci 36:653–662. https://doi.org/10.1016/j.tibs.2011.08.003
Schow EV, Freites JA, Cheng P et al (2011) Arginine in membranes: the connection between molecular dynamics simulations and translocon-mediated insertion experiments. J Membr Biol 239:35–48. https://doi.org/10.1007/s00232-010-9330-x
Gumbart JC, Teo I, Roux B, Schulten K (2013) Reconciling the roles of kinetic and thermodynamic factors in membrane-protein insertion. J Am Chem Soc 135:2291–2297. https://doi.org/10.1021/ja310777k
Junne T, Kocik L, Spiess M (2010) The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol Biol Cell 21:1662–1670. https://doi.org/10.1091/mbc.E10-01-0060
Demirci E, Junne T, Baday S et al (2013) Functional asymmetry within the Sec61p translocon. Proc Natl Acad Sci USA 110:18856–18861. https://doi.org/10.1073/pnas.1318432110
Ismail N, Hedman R, Schiller N, von Heijne G (2012) A biphasic pulling force acts on transmembrane helices during translocon-mediated membrane integration. Nat Struct Mol Biol 19:1018–1022. https://doi.org/10.1038/nsmb.2376
Junne T, Spiess M (2017) Integration of transmembrane domains is regulated by their downstream sequences. J Cell Sci 130:372–381. https://doi.org/10.1242/jcs.194472
Lundin C, Kim H, Nilsson I et al (2008) Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc Natl Acad Sci USA 105:15702–15707. https://doi.org/10.1073/pnas.0804842105
Cymer F, Ismail N, von Heijne G (2014) Weak pulling forces exerted on Nin-orientated transmembrane segments during co-translational insertion into the inner membrane of Escherichia coli. FEBS Lett 588:1930–1934. https://doi.org/10.1016/j.febslet.2014.03.050
Deshaies RJ, Sanders SL, Feldheim DA, Schekman R (1991) Assembly of yeast Sec proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349:806–808. https://doi.org/10.1038/349806a0
Panzner S, Dreier L, Hartmann E et al (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 81:561–570
Itskanov S, Park E (2019) Structure of the posttranslational Sec protein-translocation channel complex from yeast. Science 363:84–87. https://doi.org/10.1126/science.aav6740
Wu X, Cabanos C, Rapoport TA (2019) Structure of the post-translational protein translocation machinery of the ER membrane. Nature 566:136–139. https://doi.org/10.1038/s41586-018-0856-x
Brodsky JL, Goeckeler J, Schekman R (1995) BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc Natl Acad Sci USA 92:9643–9646
Young BP, Craven RA, Reid PJ et al (2001) Sec63p and Kar2p are required for the translocation of SRP-dependent precursors into the yeast endoplasmic reticulum in vivo. EMBO J 20:262–271. https://doi.org/10.1093/emboj/20.1.262
Lang S, Benedix J, Fedeles SV et al (2012) Different effects of Sec61α, Sec62 and Sec63 depletion on transport of polypeptides into the endoplasmic reticulum of mammalian cells. J Cell Sci 125:1958–1969. https://doi.org/10.1242/jcs.096727
Voigt S, Jungnickel B, Hartmann E, Rapoport TA (1996) Signal sequence-dependent function of the TRAM protein during early phases of protein transport across the endoplasmic reticulum membrane. J Cell Biol 134:25–35
Chen Q, Denard B, Lee C-E et al (2016) Inverting the topology of a transmembrane protein by regulating the translocation of the first transmembrane helix. Mol Cell 63:567–578. https://doi.org/10.1016/j.molcel.2016.06.032
Jonikas MC, Collins SR, Denic V et al (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323:1693–1697. https://doi.org/10.1126/science.1167983
Chitwood PJ, Juszkiewicz S, Guna A et al (2018) EMC is required to initiate accurate membrane protein topogenesis. Cell 175:1507–1519.e16. https://doi.org/10.1016/j.cell.2018.10.009
Shurtleff MJ, Itzhak DN, Hussmann JA et al (2018) The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. Elife 7:382. https://doi.org/10.7554/eLife.37018
van den Berg B, Clemons WM, Collinson I et al (2004) X-ray structure of a protein-conducting channel. Nature 427:36–44. https://doi.org/10.1038/nature02218
Sommer N, Junne T, Kalies K-U et al (2013) TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim Biophys Acta 1833:3104–3111. https://doi.org/10.1016/j.bbamcr.2013.08.018
Hessa T, Reithinger JH, von Heijne G, Kim H (2009) Analysis of transmembrane helix integration in the endoplasmic reticulum in S. cerevisiae. J Mol Biol 386:1222–1228
Xie K, Hessa T, Seppälä S et al (2007) Features of transmembrane segments that promote the lateral release from the translocase into the lipid phase. Biochemistry 46:15153–15161. https://doi.org/10.1021/bi701398y
Ojemalm K, Botelho SC, Stüdle C, von Heijne G (2013) Quantitative analysis of SecYEG-mediated insertion of transmembrane α-helices into the bacterial inner membrane. J Mol Biol 425:2813–2822. https://doi.org/10.1016/j.jmb.2013.04.025
Acknowledgements
Our work was supported by Grant No. 31003A-182519 from the Swiss National Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Spiess, M., Junne, T. & Janoschke, M. Membrane Protein Integration and Topogenesis at the ER. Protein J 38, 306–316 (2019). https://doi.org/10.1007/s10930-019-09827-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-019-09827-6