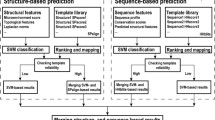
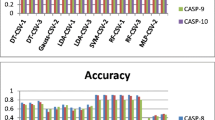
Abstract
Predicting catalytic sites of a given enzyme is an important open problem of Bioinformatics. Recently, many machine learning-based methods have been developed which have the advantage that they can account for many sequential or structural features. We found that although many kinds of features are incorporated, protein sequence conservation is the main part of information they used and should play an important role in the future. So we tested several conservation features in their ability to predict catalytic sites by using the Support Vector Machine classifier. Our results suggest that position specific scoring matrix performs better than other features and incorporating conservation information of sequentially adjacent sites is more effective than that of structurally adjacent ones. Moreover, although conservation information is effective in predicting catalytic sites, it is a difficult problem to optimize the combination of conservation features and other ones.








Similar content being viewed by others
Abbreviations
- SVM:
-
Support Vector Machine
- JSD:
-
Jensen-Shannon divergence
- PSSM:
-
Position specific scoring matrix
- PCA:
-
Principal Component Analysis
- PC:
-
Principal Component
- WOP:
-
Weighted observed percentage
- CR:
-
Contribution rate
- ACR:
-
Accumulated contribution rate
- ROC:
-
Receiver operating characteristic
- P:
-
Precision
- R:
-
Recall
- FPR:
-
False positive rate
- TPR:
-
True position rate
- MCC:
-
Matthews correlation coefficient
- RP:
-
Recall/Precision
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucleic Acids Res 25(17):3398–3402
Caffery D, Somaroo S, Hughes J, Mintserlis J, Hunang E (2004) Protein Sci 13:190–202
Capra J, Singh S (2007) Bioinformatics 23:1875–1882
Cilia E, Passerini A (2010) BMC Bioinformat 11:115
del sol mesa A, Pazos F, Valencia A (2003) J Mol Biol 326:1289–1302
Donald JS, Shakhnovich EI (2005) Bioinformatics 21:2629–2635
Dou YC, Zheng XQ, Wang J (2009) J Theor Biol 262(2):317–322
Dou YC, Zheng XQ, Wang J (2009) Protein J 28:29–33
Dou YC, Zheng XQ, Yang JL, Wang J (2010) Amino Acids 39:1353–1361
Dukka B, Dennis R (2008) Bioinformatics 24:2308–2316
Fan RE, Chen PH, Lin CJ (2005) J Mach Learn Res 6:1889–1918
Fischer JD, Mayer CE, Soding J (2008) Bioinformatics 24:613–620
Gutteridge A, Bartlett GJ, Thornton JM (2003) J Mol Biol 303:719–734
Innis CA, Anand AP, Sowdhamini R (2003) J Mol Biol 337:1053–1068
Johansson F, Toh H (2010) BMC Bioinformat 11:383
Johansson F, Toh H (2010) J Bioinform Comput Biol 8(5):809–823
Li GH, Huang JF (2010) BMC Bioinformat 11:439
Liu H, Setiono R (1995) IEEE computer society. Washington, DC, USA, pp 388–391
Liu XS, Guo WL (2008) Amino Acids 34:643–652
Liu ZP, Wu LY, Wang Y, Zhang XS, Chen LN (2010) Bioinformatics 26:1616–1622
Mayrose I, Graur D, Ben-Tal N, Pupko T (2004) Mol Biol Evol 21:1781–1791
Mihalek I, Reos I, Lichtarge O (2004) J Mol Biol 336:1265–1282
Mirny L, Shakhnovich E (1999) J Mol Biol 291:177–196
Palenchar P, Mount M, Cusato D, Dougherty J (2008) Protein J 27:401–407
Panchenko A, Kondrashov F, Bryant S (2003) Protein Sci 13:884–892
Pande S, Raheja A, Livesay DR (2007) IEEE Symp CIBCB 7:247–253
Pei J, Grishin N (2001) Bioinformatics 17:700–712
Petrova N, Wu C (2006) BMC Bioinformat 7:312
Sankararaman S, Sha F, Kirsch JF, Jordan MI, Kimmen Sjolander K (2010) Bioinformatics 5:617–624
Shenkin P, Erman BLM (1991) Proteins 11:297–313
Smith LI (2002) http://www.cs.otago.ac.nz/cosc453/student_tutorials/principal_components.pdf
Sterner B, Singh R, Berger B (2007) J Comput Biol 14:1058–1073
Tang Y, Sheng Z, Chen Y, Zhang Z (2008) Protein Eng Des Sel 21:295–302
Taylor W (1986) J Theor Biol 119:205–218
Valdar W (2002) Proteins 48:227–241
Wang K, Samudrala R (2006) BMC Bioinformat 7:385
Williamson R (1995) J Theor Biol 24:908–915
Ye K, Vriend G, IJzerman AP (2008) Bioinformatics 24:908–915
Youn E (2007) Protein Sci 16:216–226
Zhang SW, Zhang YL, Pan Q, Cheng YM, Chou KC (2008) Amino Acids 35:495–501
Zhang T, Zhang H, Chen K, Shen SY, Ruan JS, Kurgan L (2008) Bioinformatics 24:2329–2338
Acknowledgments
This work was partially supported by the National Natural Science Foundation of China (No. 10731040), Shanghai Leading Academic Discipline Project (No. S30405) and Innovation Program of Shanghai Municipal Education Commission (No. 09zz134). All the calculational tasks are applied on a LENOVO Shenteng 1800 COW, which is located in School of Mathematical Science, Dalian University of Technology. The authors thank DR. Elisa Cilia for providing their data on the HA-superfamily data set.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dou, Y., Geng, X., Gao, H. et al. Sequence Conservation in the Prediction of Catalytic Sites. Protein J 30, 229–239 (2011). https://doi.org/10.1007/s10930-011-9324-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-011-9324-2