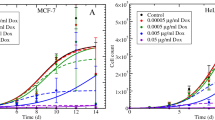
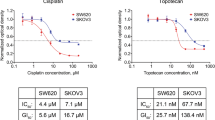
Abstract
Concentration response experiments are utilized widely to characterize the response of tumor cell lines to chemotherapeutic drugs, but the assay methods are non-standardized and their analysis based on phenomenological equations. To provide a framework for better interpretation of these experiments, we have developed a mathematical model in which progression through the tumor cell cycle is inhibited by drug treatment via either cell cycle arrest or entrance into cell death pathways. By fitting concentration response data, preferably over a dynamic range, the contributions of these mechanisms can be delineated. The model was shown to fit well experimental data for three glioma cell lines treated with either carmustine or etoposide. In each cell line, the major mechanism of tumor cell inhibition was cell death for carmustine in contrast to cell cycle arrest for etoposide. The model also provides a possible interpretation for the acquired in vitro resistance of U87 cells to carmustine as an accelerated desensitization to cell killing effects. This approach will aid in understanding better the action of chemotherapeutic agents on tumor cells.









Similar content being viewed by others
References
Basse B, Baguley BC, Marshall ES, Joseph WR, van Brunt B, Wake G, Wall DJ (2004) Modelling cell death in human tumour cell lines exposed to the anticancer drug paclitaxel. J Math Biol 49:329–357
Kozusko F, Chen P, Grant SG, Day BW, Panetta JC (2001) A mathematical model of in vitro cancer cell growth and treatment with the antimitotic agent curacin A. Math Biosci 170:1–16
Panetta JC, Evans WE, Cheok MH (2006) Mechanistic mathematical modelling of mercaptopurine effects on cell cycle of human acute lymphoblastic leukaemia cells. Br J Cancer 94:93–100
Xu GW, Nutt CL, Zlatescu MC, Keeney M, Chin-Yee I, Cairncross JG (2001) Inactivation of p53 sensitizes U87MG glioma cells to 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res 61:4155–4159
Batista LF, Roos WP, Christmann M, Menck CF, Kaina B (2007) Differential sensitivity of malignant glioma cells to methylating and chloroethylating anticancer drugs: p53 determines the switch by regulating xpc, ddb2, and DNA double-strand breaks. Cancer Res 67:11886–11895
Yan L, Donze JR, Liu L (2005) Inactivated MGMT by O6-benzylguanine is associated with prolonged G2/M arrest in cancer cells treated with BCNU. Oncogene 24:2175–2183
Das CM, Aguilera D, Vasquez H, Prasad P, Zhang M, Wolff JE, Gopalakrishnan V (2007) Valproic acid induces p21 and topoisomerase-II (alpha/beta) expression and synergistically enhances etoposide cytotoxicity in human glioblastoma cell lines. J Neurooncol 85:159–170
Branzeiand D, Foiani M (2008) Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol 9:297–308
Chiu CC, Li CH, Ung MW, Fuh TS, Chen WL, Fang K (2005) Etoposide (VP-16) elicits apoptosis following prolonged G2–M cell arrest in p53-mutated human non-small cell lung cancer cells. Cancer Lett 223:249–258
Mayo LD, Dixon JE, Durden DL, Tonks NK, Donner DB (2002) PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J Biol Chem 277:5484–5489
Xu GW, Mymryk JS, Cairncross JG (2005) Inactivation of p53 sensitizes astrocytic glioma cells to BCNU and temozolomide, but not cisplatin. J Neurooncol 74:141–149
Yin D, Tamaki N, Kokunai T (2000) Wild-type p53-dependent etoposide-induced apoptosis mediated by caspase-3 activation in human glioma cells. J Neurosurg 93:289–297
Florian JA Jr, Eiseman JL, Parker RS (2005) Accounting for quiescent cells in tumour growth and cancer treatment. Syst Biol (Stevenage) 152:185–192
Sherer E, Hannemann RE, Rundell A, Ramkrishna D (2006) Analysis of resonance chemotherapy in leukemia treatment via multi-staged population balance models. J Theor Biol 240:648–661
Ribba B, Colin T, Schnell S (2006) A multiscale mathematical model of cancer, and its use in analyzing irradiation therapies. Theor Biol Med Model 3:7
Venkatasubramanian R, Henson MA, Forbes NS (2008) Integrating cell-cycle progression, drug penetration and energy metabolism to identify improved cancer therapeutic strategies. J Theor Biol 253:98–117
Damia G, Broggini M (2004) Improving the selectivity of cancer treatments by interfering with cell response pathways. Eur J Cancer 40:2550–2559
Senderowicz AM (2004) Targeting cell cycle and apoptosis for the treatment of human malignancies. Curr Opin Cell Biol 16:670–678
Gardner SN (2000) A mechanistic, predictive model of dose-response curves for cell cycle phase-specific and -nonspecific drugs. Cancer Res 60:1417–1425
Khoshyomn S, Nathan D, Manske GC, Osler TM, Penar PL (2002) Synergistic effect of genistein and BCNU on growth inhibition and cytotoxicity of glioblastoma cells. J Neurooncol 57:193–200
Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W (2008) Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res 14:2900–2908
Law ME, Templeton KL, Kitange G, Smith J, Misra A, Feuerstein BG, Jenkins RB (2005) Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet 160:1–14
Bar EE, Chaudhry A, Lin A, Fan X, Schreck K, Matsui W, Piccirillo S, Vescovi AL, DiMeco F, Olivi A, Eberhart CG (2007) Cyclopamine-mediated hedgehog pathway inhibition depletes stem-like cancer cells in glioblastoma. Stem Cells 25:2524–2533
Iwadate Y, Fujimoto S, Namba H, Yamaura A (2003) Promising survival for patients with glioblastoma multiforme treated with individualised chemotherapy based on in vitro drug sensitivity testing. Br J Cancer 89:1896–1900
Camphausen K, Purow B, Sproull M, Scott T, Ozawa T, Deen DF, Tofilon PJ (2005) Influence of in vivo growth on human glioma cell line gene expression: convergent profiles under orthotopic conditions. Proc Natl Acad Sci USA 102:8287–8292
Ogunnaike BA (2006) Elucidating the digital control mechanism for DNA damage repair with the p53-Mdm2 system: single cell data analysis and ensemble modelling. J R Soc Interface 3:175–184
Gurkan E, Schupp JE, Aziz MA, Kinsella TJ, Loparo KA (2007) Probabilistic modeling of DNA mismatch repair effects on cell cycle dynamics and iododeoxyuridine-DNA incorporation. Cancer Res 67:10993–11000
Haberichter T, Madge B, Christopher RA, Yoshioka N, Dhiman A, Miller R, Gendelman R, Aksenov SV, Khalil IG, Dowdy SF (2007) A systems biology dynamical model of mammalian G1 cell cycle progression. Mol Syst Biol 3:84
Ma L, Wagner J, Rice JJ, Hu W, Levine AJ, Stolovitzky GA (2005) A plausible model for the digital response of p53 to DNA damage. Proc Natl Acad Sci USA 102:14266–14271
Toettcher JE, Loewer A, Ostheimer GJ, Yaffe MB, Tidor B, Lahav G (2009) Distinct mechanisms act in concert to mediate cell cycle arrest. Proc Natl Acad Sci USA 106:785–790
Acknowledgments
Funding support was provided by the Charles and Johanna Busch Memorial Fund and an NSF IGERT Fellowship (DGE 0333196) to SH. We thank Debrabata Banerjee for advice on generating resistance and Ioannis Androulakis for a discussion on parameter estimation. We thank Theresa Choi at the Flow Cytometry Lab at EOHSI for help with ModFit for cell cycle phase quantitation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hamed, S.S., Roth, C.M. Mathematical modeling to distinguish cell cycle arrest and cell killing in chemotherapeutic concentration response curves. J Pharmacokinet Pharmacodyn 38, 385–403 (2011). https://doi.org/10.1007/s10928-011-9199-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-011-9199-z