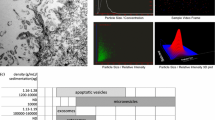
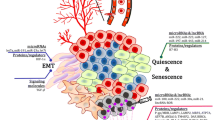
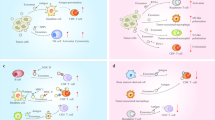
Abstract
Breast cancer (BC) is responsible for 15% of all the cancer deaths among women in the USA. The tumor microenvironment (TME) has the potential to act as a driver of breast cancer progression and metastasis. The TME is composed of stromal cells within an extracellular matrix and soluble cytokines, chemokines and extracellular vesicles and nanoparticles that actively influence cell behavior. Extracellular vesicles include exosomes, microvesicles and large oncosomes that orchestrate fundamental processes during tumor progression through direct interaction with target cells. Long before tumor cell spread to future metastatic sites, tumor-secreted exosomes enter the circulation and establish distant pre-metastatic niches, hospitable and permissive milieus for metastatic colonization. Emerging evidence suggests that breast cancer exosomes promote tumor progression and metastasis by inducing vascular leakiness, angiogenesis, invasion, immunomodulation and chemoresistance. Exosomes are found in almost all physiological fluids including plasma, urine, saliva, and breast milk, providing a valuable resource for the development of non-invasive cancer biomarkers. Here, we review work on the role of exosomes in breast cancer progression and metastasis, and describe the most recent advances in models of exosome secretion, isolation, characterization and functional analysis. We highlight the potential applications of plasma-derived exosomes as predictive biomarkers for breast cancer diagnosis, prognosis and therapy monitoring. We finally describe the therapeutic approaches of exosomes in breast cancer.


Similar content being viewed by others
Abbreviations
- AF4:
-
Asymmetric flow field-flow fractionation
- AFM:
-
Atomic force microscopy
- BC:
-
Breast cancer
- DC:
-
Dendritic cell
- DCIS:
-
Ductal carcinoma in situ
- ELISA:
-
Enzyme-linked immunosorbent assay
- EV:
-
Extracellular vesicle
- HMEC:
-
Human mammary epithelial cells
- K:
-
Cytokeratin
- MS:
-
Mass spectrometry
- MV:
-
Microvesicle
- NACT:
-
Neoadjuvant chemotherapy
- NK:
-
Natural killer
- NTA:
-
Nanoparticle tracking analysis
- PMN:
-
Pre-metastatic niche
- TDLU:
-
Terminal ductal lobular unit
- TEM:
-
Transmission electron microscopy
- TME:
-
Tumor microenvironment
- WB:
-
Western blot
References
Feng Y, et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5(2):77–106.
Radisky DC, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436(7047):123–7.
Gudjonsson T, et al. Normal and tumor-derived myoepithelial cells differ in their ability to interact with luminal breast epithelial cells for polarity and basement membrane deposition. J Cell Sci. 2002;115(Pt 1):39–50.
Bhowmick NA, et al. TGF-beta signaling in fibroblasts modulates the oncogenic potential of adjacent epithelia. Science. 2004;303(5659):848–51.
Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14(5):518–27.
Ingthorsson S, et al. Endothelial cells stimulate growth of normal and cancerous breast epithelial cells in 3D culture. BMC Res Notes. 2010;3:184.
Kraman M, et al. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330(6005):827–30.
Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9(4):285–93.
Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35.
Wortzel I, et al. Exosome-Mediated Metastasis: Communication from a Distance. Dev Cell. 2019;49(3):347–60.
Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19(2):43–51.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science, 2020. 367(6478).
Mathieu M, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17.
Skotland T, et al. An emerging focus on lipids in extracellular vesicles. Adv Drug Deliv Rev, 2020.
van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–28.
Zhang H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. 2018;20(3):332–43.
Fitzgerald W, et al. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci Rep. 2018;8(1):8973.
Jeppesen DK, et al. Reassessment of exosome composition. Cell, 2019. 177(2): p. 428–445 e18.
Ratajczak J, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20(5):847–56.
Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24(6):766–9.
Sansone P, et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc Natl Acad Sci U S A. 2017;114(43):E9066–75.
Konoshenko MY, et al. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res Int. 2018;2018:8545347.
Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–91.
Witwer KW, et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles. 2013;2:20360.
Linares R, et al. High-speed centrifugation induces aggregation of extracellular vesicles. J Extracell Vesicles. 2015;4:29509.
Sodar BW, et al. Low-density lipoprotein mimics blood plasma-derived exosomes and microvesicles during isolation and detection. Sci Rep. 2016;6:24316.
Gupta S, et al. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther. 2018;9(1):180.
Zhang Q, et al. Transfer of functional cargo in exomeres. Cell Rep, 2019. 27(3): p. 940–954 e6.
van der Pol E, et al. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8(12):2596–607.
Raposo G, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med. 1996;183(3):1161–72.
Arraud N, et al. Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J Thromb Haemost. 2014;12(5):614–27.
Skliar M, Chernyshev VS. Imaging of extracellular vesicles by atomic force microscopy. J Vis Exp, 2019(151).
Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7(6):780–8.
Anderson W, et al. Observations of Tunable Resistive Pulse Sensing for Exosome Analysis: Improving System Sensitivity and Stability. Langmuir. 2015;31(23):6577–87.
Kesimer M, Gupta R. Physical characterization and profiling of airway epithelial derived exosomes using light scattering. Methods. 2015;87:59–63.
Zheng Y, et al. Monitoring the Rab27 associated exosome pathway using nanoparticle tracking analysis. Exp Cell Res. 2013;319(12):1706–13.
Soo CY, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136(2):192–7.
Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015;17(6):816–26.
Chiang CY, Chen C. Toward characterizing extracellular vesicles at a single-particle level. J Biomed Sci. 2019;26(1):9.
Steen HB. Flow cytometer for measurement of the light scattering of viral and other submicroscopic particles. Cytometry A. 2004;57(2):94–9.
Hercher M, Mueller W, Shapiro HM. Detection and discrimination of individual viruses by flow cytometry. J Histochem Cytochem. 1979;27(1):350–2.
Yang L, et al. Development of an ultrasensitive dual-channel flow cytometer for the individual analysis of nanosized particles and biomolecules. Anal Chem. 2009;81(7):2555–63.
Lacroix R, et al. Overcoming limitations of microparticle measurement by flow cytometry. Semin Thromb Hemost. 2010;36(8):807–18.
van der Vlist EJ, et al. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat Protoc. 2012;7(7):1311–26.
Pospichalova V, et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J Extracell Vesicles. 2015;4:25530.
Morales-Kastresana A, et al. Labeling Extracellular Vesicles for Nanoscale Flow Cytometry. Sci Rep. 2017;7(1):1878.
van der Pol E, et al. Absolute sizing and label-free identification of extracellular vesicles by flow cytometry. Nanomedicine. 2018;14(3):801–10.
Tamkovich SN, et al. Isolation and characterization of exosomes from blood plasma of breast cancer and colorectal cancer patients. Biomed Khim. 2017;63(2):165–9.
Choi DS, et al. Proteomics of extracellular vesicles: Exosomes and ectosomes. Mass Spectrom Rev. 2015;34(4):474–90.
Rontogianni S, et al. Proteomic profiling of extracellular vesicles allows for human breast cancer subtyping. Commun Biol. 2019;2:325.
Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289(7):3869–75.
Pegtel DM, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci USA. 2010;107(14):6328–33.
Kosaka N, et al. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285(23):17442–52.
Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–44.
Prendergast EN, et al. Optimizing exosomal RNA isolation for RNA-Seq analyses of archival sera specimens. PLoS ONE. 2018;13(5):e0196913.
Tang YT, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40(3):834–44.
Johnsen KB, et al. What is the blood concentration of extracellular vesicles? Implications for the use of extracellular vesicles as blood-borne biomarkers of cancer. Biochim Biophys Acta Rev Cancer. 2019;1871(1):109–16.
Ando W, et al. Novel breast cancer screening: combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci Rep. 2019;9(1):13595.
Street JM, et al. Urine Exosome Isolation and Characterization. Methods Mol Biol. 2017;1641:413–23.
Gheinani AH, et al. Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery. Sci Rep. 2018;8(1):3945.
Kang Y, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3(6):537–49.
Gupta GP, et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol. 2005;70:149–58.
Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436(7050):518–24.
Bos PD, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–9.
Li J, et al. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J Extracell Vesicles. 2015;4:26883.
Labarge MA, Garbe JC, Stampfer MR. Processing of human reduction mammoplasty and mastectomy tissues for cell culture. J Vis Exp. 2013;71:e50011.
Briem E, et al. Application of the D492 Cell Lines to Explore Breast Morphogenesis, EMT and Cancer Progression in 3D Culture. J Mammary Gland Biol Neoplasia. 2019;24(2):139–47.
Garbe JC, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69(19):7557–68.
Lee JK, et al. Different culture media modulate growth, heterogeneity, and senescence in human mammary epithelial cell cultures. PLoS ONE. 2018;13(10):e0204645.
Garbe JC, et al. Immortalization of normal human mammary epithelial cells in two steps by direct targeting of senescence barriers does not require gross genomic alterations. Cell Cycle. 2014;13(21):3423–35.
Chanson L, et al. Self-organization is a dynamic and lineage-intrinsic property of mammary epithelial cells. Proc Natl Acad Sci USA. 2011;108(8):3264–9.
LaBarge MA, et al. Breast Cancer beyond the Age of Mutation. Gerontology. 2016;62(4):434–42.
Lin CH, et al. Microenvironment rigidity modulates responses to the HER2 receptor tyrosine kinase inhibitor lapatinib via YAP and TAZ transcription factors. Mol Biol Cell. 2015;26(22):3946–53.
Pelissier Vatter FA, et al. High-Dimensional Phenotyping Identifies Age-Emergent Cells in Human Mammary Epithelia. Cell Rep. 2018;23(4):1205–19.
Pelissier FA, et al. Age-related dysfunction in mechanotransduction impairs differentiation of human mammary epithelial progenitors. Cell Rep. 2014;7(6):1926–39.
Garbe JC, et al. Accumulation of multipotent progenitors with a basal differentiation bias during aging of human mammary epithelia. Cancer Res. 2012;72(14):3687–701.
Chin AR, et al. Polarized Secretion of Extracellular Vesicles by Mammary Epithelia. J Mammary Gland Biol Neoplasia. 2018;23(3):165–76.
Dutta S, et al. Interactions between exosomes from breast cancer cells and primary mammary epithelial cells leads to generation of reactive oxygen species which induce DNA damage response, stabilization of p53 and autophagy in epithelial cells. PLoS ONE. 2014;9(5):e97580.
Riches A, et al. Regulation of exosome release from mammary epithelial and breast cancer cells - a new regulatory pathway. Eur J Cancer. 2014;50(5):1025–34.
Gonzalez E, et al. Human mammospheres secrete hormone-regulated active extracellular vesicles. PLoS ONE. 2014;9(1):e83955.
Rosenbluth JM, et al. Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat Commun. 2020;11(1):1711.
Admyre C, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007;179(3):1969–78.
Pisano C, et al. Human Breast Milk-Derived Extracellular Vesicles in the Protection Against Experimental Necrotizing Enterocolitis. J Pediatr Surg. 2020;55(1):54–8.
Liao Y, et al. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res. 2017;61(11):1700082.
Yang Y, et al. Immunocompetent mouse allograft models for development of therapies to target breast cancer metastasis. Oncotarget. 2017;8(19):30621–43.
Fantozzi A, Christofori G. Mouse models of breast cancer metastasis. Breast Cancer Res. 2006;8(4):212.
Tsai PP, et al. Effects of different blood collection methods on indicators of welfare in mice. Lab Anim (NY). 2015;44(8):301–10.
Thompson C, Keck K, and Hielscher A. Isolation of Intact, Whole Mouse Mammary Glands for Analysis of Extracellular Matrix Expression and Gland Morphology. J Vis Exp. 2017(128).
Smalley MJ. Isolation, culture and analysis of mouse mammary epithelial cells. Methods Mol Biol. 2010;633:139–70.
Deugnier MA, et al. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dbeta cell line. Dev Biol. 2006;293(2):414–25.
DePeters EJ, Hovey RC. Methods for collecting milk from mice. J Mammary Gland Biol Neoplasia. 2009;14(4):397–400.
Yu H, Mouw JK, Weaver VM. Forcing form and function: biomechanical regulation of tumor evolution. Trends Cell Biol. 2011;21(1):47–56.
Northey JJ, Przybyla L, Weaver VM. Tissue Force Programs Cell Fate and Tumor Aggression. Cancer Discov. 2017;7(11):1224–37.
Bissell MJ, et al. The organizing principle: microenvironmental influences in the normal and malignant breast. Differentiation. 2002;70(9–10):537–46.
Lee GY, et al. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat Methods. 2007;4(4):359–65.
Suetsugu A, et al. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65(3):383–90.
Koumangoye RB, et al. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE. 2011;6(9):e24234.
Rodrigues G, et al. Tumour exosomal CEMIP protein promotes cancer cell colonization in brain metastasis. Nat Cell Biol. 2019;21(11):1403–12.
Wiklander OP, et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J Extracell Vesicles. 2015;4:26316.
Rivoltini L, et al. TNF-Related Apoptosis-Inducing Ligand (TRAIL)-Armed Exosomes Deliver Proapoptotic Signals to Tumor Site. Clin Cancer Res. 2016;22(14):3499–512.
Zhu L, et al. Novel alternatives to extracellular vesicle-based immunotherapy - exosome mimetics derived from natural killer cells. Artif Cells Nanomed Biotechnol. 2018;46(sup3):S166–79.
Wen SW, et al. The Biodistribution and Immune Suppressive Effects of Breast Cancer-Derived Exosomes. Cancer Res. 2016;76(23):6816–27.
Rashid MH, et al. Differential in vivo biodistribution of (131)I-labeled exosomes from diverse cellular origins and its implication for theranostic application. Nanomedicine. 2019;21:102072.
O’Brien K, et al. Exosomes from triple-negative breast cancer cells can transfer phenotypic traits representing their cells of origin to secondary cells. Eur J Cancer. 2013;49(8):1845–59.
Cho JA, et al. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int J Oncol. 2012;40(1):130–8.
Galindo-Hernandez O, et al. Extracellular vesicles from women with breast cancer promote an epithelial-mesenchymal transition-like process in mammary epithelial cells MCF10A. Tumour Biol. 2015;36(12):9649–59.
Melo SA, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–21.
Le MT, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124(12):5109–28.
Antonyak MA, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A. 2011;108(12):4852–7.
Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25(4):501–15.
Maji S, et al. Exosomal Annexin II Promotes Angiogenesis and Breast Cancer Metastasis. Mol Cancer Res. 2017;15(1):93–105.
Othman N, Jamal R, Abu N. Cancer-Derived Exosomes as Effectors of Key Inflammation-Related Players. Front Immunol. 2019;10:2103.
Clayton A, Tabi Z. Exosomes and the MICA-NKG2D system in cancer. Blood Cells Mol Dis. 2005;34(3):206–13.
Yu S, et al. Tumor exosomes inhibit differentiation of bone marrow dendritic cells. J Immunol. 2007;178(11):6867–75.
Liu C, et al. Murine mammary carcinoma exosomes promote tumor growth by suppression of NK cell function. J Immunol. 2006;176(3):1375–85.
Chow A, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Sci Rep. 2014;4:5750.
de Visser KE, Eichten A, Coussens LM. Paradoxical roles of the immune system during cancer development. Nat Rev Cancer. 2006;6(1):24–37.
Yang M, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117.
Rupp AK, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122(2):437–46.
Khan S, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer. 2014;14:176.
Galindo-Hernandez O, et al. Elevated concentration of microvesicles isolated from peripheral blood in breast cancer patients. Arch Med Res. 2013;44(3):208–14.
Kibria G, et al. A rapid, automated surface protein profiling of single circulating exosomes in human blood. Sci Rep. 2016;6:36502.
Chen IH, et al. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc Natl Acad Sci U S A. 2017;114(12):3175–80.
Hoshino A, et al. Extracellular vesicle and particle biomarkers define multiple human cancers.Cell, 2020;182(4): p. 1044–1061 e18.
Konig L, et al. Elevated levels of extracellular vesicles are associated with therapy failure and disease progression in breast cancer patients undergoing neoadjuvant chemotherapy. Oncoimmunology. 2017;7(1):e1376153.
Keklikoglou I, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol. 2019;21(2):190–202.
Chen Y, et al. Breast cancer resistance protein (BCRP)-containing circulating microvesicles contribute to chemoresistance in breast cancer. Oncol Lett. 2015;10(6):3742–8.
Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292.
Vigorito E, et al. miR-155: an ancient regulator of the immune system. Immunol Rev. 2013;253(1):146–57.
Wei Y, et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation. 2013;127(15):1609–19.
Delcayre A, et al. Exosome Display technology: applications to the development of new diagnostics and therapeutics. Blood Cells Mol Dis. 2005;35(2):158–68.
Tian Y, et al. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials. 2014;35(7):2383–90.
Wang L, et al. Exosomal pMHC-I complex targets T cell-based vaccine to directly stimulate CTL responses leading to antitumor immunity in transgenic FVBneuN and HLA-A2/HER2 mice and eradicating trastuzumab-resistant tumor in athymic nude mice. Breast Cancer Res Treat. 2013;140(2):273–84.
Escudier B, et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: results of thefirst phase I clinical trial. J Transl Med. 2005;3(1):10.
Morse MA, et al. A phase I study of dexosome immunotherapy in patients with advanced non-small cell lung cancer. J Transl Med. 2005;3(1):9.
Besse B, et al. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5(4):e1071008.
Chen YS, et al. Exosomes in clinical trial and their production in compliance with good manufacturing practice. Ci Ji Yi Xue Za Zhi. 2019;32(2):113–20.
Wong NC, et al. Alphav integrins mediate adhesion and migration of breast carcinoma cell lines. Clin Exp Metastasis. 1998;16(1):50–61.
Jang JY, et al. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13:421.
Ohno S, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21(1):185–91.
Thery C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7(1):1535750.
Acknowledgments
The authors gratefully acknowledge support from Dr. David Lyden.
The figures were created using Servier Medical Art templates, which are licensed under a Creative Commons Attribution 3.0 Unported License; https://smart.servier.com. F.A.V was supported by a Swiss National Science Foundation (SFNS) Postdoc.Mobility grant (P2SKP3_174785 and P400PB_186791). S.L was supported by United States Department of Defense W81XWH2010263-01. H.Z was supported by National Cancer Institute CA218513 and U19 AI144301-01.
Author information
Authors and Affiliations
Contributions
The first draft of the manuscript was written by Fanny A. Pelissier Vatter who performed the literature search and data analysis. Serena Lucotti and Haiying Zhang critically revised the work.
Corresponding author
Ethics declarations
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pelissier Vatter, F.A., Lucotti, S. & Zhang, H. Recent Advances in Experimental Models of Breast Cancer Exosome Secretion, Characterization and Function. J Mammary Gland Biol Neoplasia 25, 305–317 (2020). https://doi.org/10.1007/s10911-020-09473-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-020-09473-0