Abstract
In order to properly understand the spread of SARS-CoV-2 infection and development of humoral immunity, researchers have evaluated the presence of serum antibodies of people worldwide experiencing the pandemic. These studies rely on the use of recombinant proteins from the viral genome in order to identify serum antibodies that recognize SARS-CoV-2 epitopes. Here, we discuss the cross-reactivity potential of SARS-CoV-2 antibodies with the full spike proteins of four other betacoronaviruses that cause disease in humans, MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1. Using enzyme-linked immunosorbent assays (ELISAs), we detected the potential cross-reactivity of antibodies against SARS-CoV-2 towards the four other coronaviruses, with the strongest cross-recognition between SARS-CoV-2 and SARS /MERS-CoV antibodies, as expected based on sequence homology of their respective spike proteins. Further analysis of cross-reactivity could provide informative data that could lead to intelligently designed pan-coronavirus therapeutics or vaccines.
Similar content being viewed by others
Introduction
The SARS-CoV-2 pandemic has reached almost every country on Earth. As with many viral infections, our immune system responds to SARS-CoV-2 infection through a variety of cellular and humoral effectors. These include antibodies produced by B cells, which can be formed against various viral proteins. For SARS-CoV-2, antibodies have been detected that recognize three of the four SARS-CoV-2 proteins exposed on the surface of the viral capsid: the nucleocapsid (N), envelope (E), and spike (S) proteins [1]. The spike protein forms as a homotrimer and mediates receptor binding through its receptor binding domain (RBD) to host cell ACE2 and is thus the major target of neutralizing antibody responses [2, 3]. When testing for the presence of SARS-CoV-2 antibodies, researchers have utilized the full spike ectodomain as well as the RBD domain alone for antigens in enzyme-linked immunosorbent assays (ELISAs) and other serologic assays [4].
The zoonotic betacoronaviruses SARS-CoV and SARS-CoV-2 (endemic/pandemic sarbecoviruses), and MERS (endemic merbecovirus) derived primarily from wildlife, while the viruses HCoV-OC43 and HCoV-HKU1 (seasonal embecoviruses) are endemic in humans [5, 6]. All of these viruses bear the spike protein on their surface [7, 8]. As such, anti-spike antibodies are common in response to each of the five human-infecting betacoronaviruses [9,10,11]. Knowledge of cross-reactivity of anti-spike antibodies against different viruses is critical for understanding of SARS-CoV-2 immunity of individuals who have had prior exposure to other betacoronaviruses and of potential future immunity of COVID-19 survivors to other coronaviruses [12]. Furthermore, knowledge of cross-reactivity is necessary to understand and properly interpret results from serologic studies such as serosurveys and clinical antibody tests [13, 14]. Previous research has shown minimal cross-reactivity between RBD domains from differing coronaviruses; however, these studies largely ignore the rest of the spike protein, which will be an important consideration for identification of potential therapeutic antibodies and can be used in vitro to help identify polyclonal responses that are not detected with RBD alone [15]. The surface of the spike protein is very large and non-RBD domains of the protein are responsible for stabilizing the prefusion configuration of the protein as well as generating the postfusion structure needed for cellular entry. Monoclonal antibodies which target these non-RBD domains of the protein have already been identified and understanding the potential cross-reactivity issues with other regions of the spike protein are essential [16].
Here, we evaluated the serologic reactivity of pre-pandemic archival blood serum samples (pre-2019, collected from healthy donors in Bethesda from 2014 to 2018) and samples collected in April 2020 from a community (collected from blood donors in the New York/New Jersey region, 62 SARS-CoV-2 seropositive, 22 known PCR+, all known close contact with PCR+ patient) highly affected by SARS-CoV-2. Utilizing 12 previously reported ELISAs [15], we tested IgG, IgM, and IgA reactivity against spike proteins from SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1.
Results
Sequence Homology Between Pandemic, Endemic, and Seasonal Coronaviruses
To evaluate the potential for cross-reactivity, we first compared the spike protein sequence homology among SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1 (Fig. 1, Supplementary Fig. 1). The greatest homology was between SARS-CoV-2 and SARS-CoV (76% identity, 87% similarity), followed by MERS-CoV (42% identity, 58% similarity) and lastly HCoV-OC43/HCoV-HKU1 (OC43 30% identity, 41% similarity; HKU1 29% identity, 40% similarity). Embecoviruses HCoV-OC43 and HCoV-HKU1 are more similar to each other (64% identity, 75% similarity) than to the two endemic betacoronaviruses. There is a larger fraction of homology towards the C-terminus of the protein in all coronavirus spike proteins, which represents the major structural regions of the protein including the heptad repeat regions responsible for insertion of the fusion peptide into the host cell membrane. Homology is significantly lower in the N-terminal regions of spike, with significant lack of similarity in the regions including the receptor-binding domain, correlating with the difference in receptors and determinants used for host cell entry in the different betacoronaviruses (MERS-CoV: receptor dipeptidyl peptidase-4 (DPP4), SARS-CoV/SAR-CoV-2: ACE2, HCoV-OC43/HCoV-HKU1: the sugar N-acetylneuraminic acid) [8].
Sequence homology of SARS-CoV-2S glycoprotein ectodomain with endemic and seasonal betacoronaviruses. SARS-CoV-2 stabilized spike protein ectodomain sequence aligned with MERS-CoV (MERS), SARS-CoV (SARS1), HCoV-OC43, and HCoV-HKU1 proteinsbetacoronaviruses. a Percent (%) similarity to SARS-CoV-2. b Percent (%) identity to SARS-CoV-2. Domain abbreviations are: NTD, N-terminal domain; RBD, receptor-binding domain; S1/S2, furin cleavage site; FP, fusion peptide; HR1/HR2, heptad repeat regions
Serologic Reactivity of Anti-Spike IgG, IgM, and IgA Antibodies
Functional cross-reactivity was determined through the use of enzyme-linked immunosorbent assays (ELISAs) measuring IgG, IgM, and IgA subclasses, representing mature, early stage, and mucosal specific serologic responses, respectively. We produced recombinant soluble spike proteins of SARS-CoV-2, MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1 using the Expi293 expression system, which yielded pure, intact ectodomain trimers suitable for ELISA [17]. Notably, the yields of all coronavirus spike proteins were significantly different even though four of five were cloned in identical vectors and contained the same modifications to the wild-type sequences (elimination of furin cleavage site, prefusion-stabilizing proline mutations (2P), similar C-terminal tags), none of which was expected to alter serologic recognition due to their internal locations. The HCoV-OC43 construct has all of these features but the wild-type furin cleavage site is present. Using similar expression conditions, SARS-CoV-2 spike was produced at a maximum of 2 mg/L culture, while the other spike proteins were significantly easier to produce with yields of 5, 11, 8, and 6 mg/L respectively for SARS-CoV, MERS-CoV, HCoV-OC43, and HCoV-HKU1.
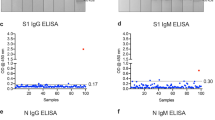
We next utilized a semi-automated ELISA protocol to detect serum antibodies from pre-2019 archival samples and samples from a community with high SARS-CoV-2 prevalence during the 2020 pandemic (Fig. 2). In serum samples collected from healthy volunteers prior to 2019, there was minimal reactivity with SARS-CoV-2, MERS-CoV and SARS-CoV. The majority of tested samples (n = 114) displayed high IgG reactivity with HCoV-OC43 and HCoV-HKU1 spike proteins, consistent with the extensive spread of seasonal betacoronavirus infections within the USA (Fig. 2a–c). As reported previously, we detected a high proportion of donors who seroconverted and were SARS-CoV-2 IgG+ in a community in New York City, along with a significant number of IgM and IgA seropositive donors, including several donors who were non-symptomatic, of these 68 donors, 62 were seropositive, and 22 had a previous known PCR+ diagnosis (Fig. 2c) [15]. All samples had low levels of IgM reactivity against MERS-CoV, SARS-CoV, HCoV-OC43, and HCoV-HKU1 (Fig. 2d–f). IgA antibodies were present at higher levels than IgM, but still well below levels of IgG, correlating well with biologic prevalence of antibody classes in response to pathogens (Fig. 2g–i).
Serologic positivity of immunoglobulins G, M, and A for five betacoronaviruses in pre-2019 and high prevalence SARS-CoV-2 blood donors. Anti-spike ELISA results for archival negative (pre-2019, black), hot-spot community symptomatic (pink), and hot-spot community asymptomatic (teal) blood donors for a–c IgG, d–f IgM, and g–i IgA. a, b Raw signal intensity for IgG. c Percent positive for IgG. d, e Raw signal intensity for IgM. f Percent positive for IgM. g, h Raw signal intensity for IgA. i Percent positive for IgA
Minimal Linear Correlation of SARS-CoV-2 Signal Intensity with Other Betacoronaviruses
When comparing the assay absorbance signal (optical density, OD) between SARS-CoV-2 and the other spike proteins in the high-incidence population, we saw a stronger correlation of signal intensity between SARS-CoV-2 and SARS-CoV IgG (Correlation = 0.711, R2 = 0.505) and the lowest correlation with HCoV-HKU1 (Correlation = 0.281, R2 = 0.079) (Fig. 3a, Supplementary Fig. 2). Though there was not a precise linear correlation for IgG, donors who represented signal intensity in the lower 50% of SARS-CoV-2 absorbance readings did have a significantly lower MERS-CoV and SARS-CoV signal intensity when compared to the upper 50% of SARS-CoV-2 intensity (Fig. 4). Overall, these data suggest some cross-reactivity occurs that is more easily detectable at high titers of antibody.
High levels of SARS-CoV-2 spike antibodies correlate with an increase in ELISA signal intensity for other betacoronavirus reactivity. Comparison of the mean absorbance (optical density, OD) of the upper (blue) and lower (red) 50% of SARS-CoV-2 signal intensity for a IgG, b IgM, and c IgA. Student’s t test with Holm-Šídák method for multiple comparisons. Alpha = 0.05. **** = p < 0.0001, *** = p < 0.001, ** = p < 0.01, * = p < 0.05
Cross-Reactivity of SARS-CoV-2 IgG Antibodies with Endemic and Seasonal Coronaviruses
Since we observed a difference in the IgG signal intensity of other betacoronaviruses with high levels of SARS-CoV-2 antibodies, we further analyzed the relationship between SARS-CoV-2 seroprevalence and antibody titer with SARS-CoV, MERS-CoV, HCoV-OC43, and HCoV-HKU1 in pre-pandemic (pre-2019), high-prevalence symptomatic donors, and high-prevalence asymptomatic donors (Fig. 5, Supplementary Fig. 3). Overall, archival pre-2019 samples displayed an equivalent low signal intensity of SARS-CoV-2, MERS-CoV, and SARS-CoV spike reactivity (Fig. 5a). One donor from this group with high reactivity for SARS-CoV-2 was negative for both MERS-CoV and SARS-CoV. As previously discussed, the majority of donors were HCoV-OC43 and HCoV-HKU1 seropositive due to the broad circulation of these viruses in humans. In the high SARS-CoV-2 incidence community, for both symptomatic and asymptomatic individuals, there appeared to be a correlation in SARS-CoV-2 signal intensity with MERS-CoV and SARS-CoV. To further analyze this, we directly compared the signal intensity of archival sample controls to the high-incidence pandemic population (Fig. 5b). There was a significant difference in signal intensity of MERS-CoV (p < 0.0001), SARS-CoV (p < 0.0001), HCoV-OC43 (p = 0.0123), and HCoV-HKU1 (p < 0.0001), suggesting potential cross-reactivity of SARS-CoV-2 IgG antibodies with all four spike proteins.
Anti-spike IgG signal intensity in SARS-CoV-2 seropositive and seronegative blood samples. a Relationship of SARS-CoV-2 spike IgG signal intensity in archival (black), symptomatic high exposure (pink) and asymptomatic high exposure (teal) donors. b Comparison of archival sample IgG reactivity with symptomatic high exposure sample reactivity. Unpaired two-tailed t test
Discussion
Cross-reactivity of antibodies with multiple coronaviruses is an important consideration in studying the SARS-CoV-2 pandemic, both technically, for identifying individuals who have been exposed to and recovered from the virus, as well as therapeutically, to identify broadly neutralizing antibodies or epitopes on multiple coronavirus subtypes [12, 18, 19]. Accordingly, we analyzed potential serologic cross-reactivity of antibodies with spike proteins derived from SARS-CoV-2 as well as two endemic (MERS-CoV, SARS-CoV) and two seasonal (HCoV-OC43, HCoV-HKU1) betacoronavirus species. Although two alphacoronaviruses also cause seasonal illness throughout the world (HCoV-229E and HCoV-NL43), the lack of significant sequence identity between the alphacoronavirus spike proteins and their betacoronavirus cousins (< 25%) suggests that they would be unlikely to have significant cross-reactivity. It is unclear, in terms of plasmid-based protein expression, why there is so much variability in spike protein expression levels between the different viruses, but this argues again for significant differences in the behavior of these proteins regardless of their primary sequence homology.
Antibodies that react with the spike proteins of HCoV-OC43 and HCoV-HKU1 are highly prevalent in the general population of the USA as determined by their measurement in archival pre-2019 serum samples. Previous reports of their prevalence show that the majority of children are exposed to HCoV-OC43 and seroconvert early in life [20]. The detection of high serologic reactivity of archival controls with HCoV-HKU1 might, thus, be due to the strong seroprevalence of HCoV-OC43 antibodies. Further studies would be needed to determine this interaction, though due to the high level of sequence and structural homology of their spike proteins, such a cross-reactivity between the two tested seasonal betacoronaviruses would not be surprising.
When compared to reactivity with the SARS-CoV-2 spike protein, antibodies that react to HCoV-OC43 and HCoV-HKU1 have minimal cross-reactivity with the pandemic SARS-CoV-2 or two other endemic coronaviruses, MERS-CoV and SARS-CoV. This phenotype correlates with the sequence homology of these proteins, wherein SARS-CoV-2 spike is more similar to SARS-CoV and MERS-CoV, as opposed to HCoV-OC43 and HCoV-HKU1 seasonal coronaviruses.
When comparing serum from healthy volunteers collected pre-2019 (archival controls) to those from a high-exposure community, we observe that SARS-CoV-2 antibodies react intermediately with MERS-CoV and SARS-CoV spike proteins. The mean ELISA signal intensity is significantly greater for both MERS-CoV and SARS-CoV when comparing archival controls versus the high-incidence community. Although there is minimal linear correlation between signal intensity of SARS-CoV-2 and MERS-CoV/SARS-CoV, the higher titer SARS-CoV-2 donors also display a significantly higher MERS-CoV and SARS-CoV signal intensity compared to their lower titer counterparts within the same population. There are previously mentioned limitations to the studies presented, and further research will be critical in fully understanding these phenomena, including studies on deep sequencing of viral genomes of confirmed PCR-positive cases in correlation with antibody cross-reactivity. This would be further improved by a study with detained travel information as well as identification of patients with known prior SARS-CoV-1, MERS-CoV, HCoV-OC43, or HCoV-HKU1 infections. Generation of hybrid coronavirus spike proteins to generate broadly neutralizing antibodies may prove fruitful in pursuit of a pan-coronavirus vaccine and will lend more information on how our immune systems respond to different viruses with similar structure.
Given the low seroprevalence of SARS-CoV and MERS-CoV outside of their endemic regions, and the significantly lower reactivity of SARS-CoV-2 patient sera to SARS-CoV and MERS-CoV spike proteins, it is likely that any reactivity between the pandemic SARS-CoV-2 pandemic and MERS-CoV/SARS-CoV endemic viruses would result in minimal noise between SARS-CoV-2 signal and endemic coronavirus signal in serological assays. In countries with a higher prevalence of MERS-CoV and SARS-CoV, researchers should include thorough analysis of archival patient sera (pre-2019), including sera from known SARS-CoV and MERS-CoV convalescent patients, to properly analyze the resulting data and adjust any estimates of seropositivity as needed. No clinical serology studies of SARS-CoV-2 immunity in populations previously infected with either SARS-CoV or MERS-CoV have yet emerged. A longitudinal serosurvey currently underway by the team authors will allow the team to examine whether cross-reactivities persist through longitudinally collected serum samples, and to confirm that the emergence of new SARS-CoV-2 strains that harbor spike protein variants will result in changes in cross-reactivity [21]. We recently demonstrated that the D614G spike variant does not alter IgG, IgM, or IgA spike seroassay performance and cross-reactivity, but if (or rather, as) drug-resistant or vaccine-escape variants appear, vigilance in confirming seroassay performance and cross-reactivity is required [22]. It is possible that in acutely ill individuals, many non-specific antibodies are produced that may react with the Spike ELISAs, but we have previously shown that no cross reactivity was observed with anti-influenza A H1N1 (unrelated viral antigen) [15].
Additionally, individuals who have strongly seroconverted after SARS-CoV-2 infection, and who display cross-reactivity for both MERS-CoV and SARS-CoV spike proteins, are of great interest for translational study. These individuals could potentially harbor antibodies that are universally reactive to multiple betacoronaviruses and, if these antibodies are functional for neutralization, could be important to identify to inform the development of novel therapeutics or vaccines.
Materials and Methods
Human Serum Samples
Archival (pre-2019) serum samples (n = 114) were collected between January 2014 and December 2018 from healthy adults (aged 18–55 years) through an existing NIH study NCT01386424. High-incidence community samples are deidentified uncoded samples donated from a community blood draw from donors in New York and New Jersey in April 2020. Twenty-two of these donors had a previous SARS-CoV-2 nasopharyngeal swab PCR-based diagnosis, 46 were symptomatic but undiagnosed, and 6 were asymptomatic but had known exposure (n = 68 symptomatic, n = 6 asymptomatic). All clinical trials were conducted in accordance with the provisions of the Declaration of Helsinki and Good Clinical Practice guidelines. All clinical trial participants signed written informed consent prior to enrollment.
Plasmid Sourcing and Preparation
SARS-CoV-2, MERS-CoV, HCoV-HKU1, HCoV-OC43, and SARS-CoV spike plasmids were produced by the McLellan lab at the University of Texas at Austin and the NIAID Vaccine Research Center and prepared as previously described [17, 23, 24]. Briefly, all spike constructs contained mammalian-codon-optimized genes encoding spike ectodomains containing 2 proline mutations for stabilization, with a C-terminal T4 fibritin trimerization domain, an HRV3C protease cleavage site, an 8xHis-tag and a Twin-Strep-tag. In all constructs except for HCoV-OC43, the S1/S2 furin-recognition site was mutated to produce a single-chain S protein. Final synthetic clones were subcloned into the eukaryotic expression vector pαH.
Protein Production and Purification
Soluble spike trimers were produced by expression in Expi293 cells and purified by a combination of tangential flow filtration, immobilized metal affinity chromatography, and desalting, following the procedures noted in Esposito et al. [17] Expression was carried out at 32 °C for 96 h prior to harvest. Final purified proteins were validated by a combination of SDS-PAGE and analytical size exclusion chromatography (AnSEC). All spike proteins produced single peaks on AnSEC over a Superdex 200 column, and the peak elution was consistent with the size of a trimeric spike protein. Of note, the HCoV-OC43 spike protein undergoes cleavage during SDS-PAGE leading to the appearance of two bands at 80 and 100 kDa as well as the appropriately full-length band migrating at 180 kDa. AnSEC confirms that this is an artifact of the SDS-PAGE process, as the protein elutes in a single trimeric peak of the appropriate size.
Enzyme-Linked Immunosorbent Assays
We performed ELISAs as previously described [15]. Briefly, spike proteins were suspended at 1 μg/ml in 1 × PBS. One hundred microliters of protein suspension was added to each well of a 96-well Nunc MaxiSorp ELISA plate and allowed to coat overnight at 4 °C for 16 h. Wells were washed three times with 300 μl of 1× PBS + 0.05% Tween20 (wash buffer) followed by blocking for 2 h at room temperature with 200 μl of 1× PBS + 0.05% Tween20 + 5% Non-fat dry milk (blocking buffer). Wells were washed again three times with 300 μl of wash buffer prior to addition of 100 μl of sample diluted in blocking buffer (serum samples were heat inactivated for 45 min at 56 °C and diluted at 1:400 in blocking buffer). Samples were incubated for 1 h at room temperature, then washed three times with 300 μl of wash buffer. One hundred microliters of 1-Step Ultra TMB Substrate (ThermoFisher) was added and the plate was incubated for 10 min prior to stopping the reaction with 1 N sulfuric acid (Stop Solution, ThermoFisher). Absorbance was read at 450 nm and 650 nm on a BioTek Epoch2 plate reader. The process is semi-automated through the use of a BioTek EL406 plate washer/dispenser and two BioStack 4 plate stackers to minimize plate-to-plate variation and increase throughput (see Klumpp-Thomas C, Kalish H et al. 2020 for detailed automation methods) [15].
Data Analysis
Absorbance values (optical density) were collected at 450 and 650 nm. Abs650 was subtracted from Abs450 to remove background signal. Data were subsequently analyzed utilizing Microsoft Excel and GraphPad Prism. Samples were determined as positive if the OD was greater than 1 for IgG, and greater than 0.5 for IgM and IgA.
References
Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med. 2020:1–4.
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C-L, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–3.
Yuan M, Wu NC, Zhu X, Lee C-CD, So RTY, Lv H, et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368(6491):630–3.
Stadlbauer D, Amanat F, Chromikova V, Jiang K, Strohmeier S, Arunkumar GA, et al. SARS-CoV-2 Seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 2020;57(1):e100.
Gaunt ER, Hardie A, Claas EC, Simmonds P, Templeton KE. Epidemiology and clinical presentations of the four human coronaviruses 229E, HKU1, NL63, and OC43 detected over 3 years using a novel multiplex real-time PCR method. J Clin Microbiol. 2010;48(8):2940–7.
Corman VM, Muth D, Niemeyer D, Drosten C. Hosts and sources of endemic human coronaviruses. Adv Virus Res. 2018;100: Elsevier:163–88.
Bosch BJ, Van der Zee R, De Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–11.
Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3:237–61.
Xia S, Yan L, Xu W, Agrawal AS, Algaissi A, Tseng C-TK, et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580.
Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S. The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol. 2009;7(3):226–36.
Gorse GJ, Patel GB, Vitale JN, O'Connor TZ. Prevalence of antibodies to four human coronaviruses is lower in nasal secretions than in serum. Clin Vaccine Immunol. 2010;17(12):1875–80.
Patrick DM, Petric M, Skowronski DM, Guasparini R, Booth TF, Krajden M, et al. An outbreak of human coronavirus OC43 infection and serological cross-reactivity with SARS coronavirus. Can J Infect Dis Med Microbiol. 2006;17:330–6.
Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:6.
Petherick A. Developing antibody tests for SARS-CoV-2. Lancet. 2020;395(10230):1101–2.
Klumpp-Thomas C, Kalish H, Drew M, Hunsberger S, Snead K, Fay MP, et al. Standardization of ELISA protocols for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. Nat Commun. 2021;12(1):113.
Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus ADME, et al. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun. 2020;11(1):2251.
Esposito D, Mehalko J, Drew M, Snead K, Wall V, Taylor T, et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr Purif. 2020;105686.
Khan S, Nakajima R, Jain A, De Assis RR, Jasinskas A, Obiero JM, et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. BioRxiv. 2020.
Huang AT, Garcia-Carreras B, Hitchings MD, Yang B, Katzelnick LC, Rattigan SM, et al. 2020, A systematic review of antibody mediated immunity to coronaviruses: antibody kinetics, correlates of protection, and association of antibody responses with severity of disease. medRxiv.
Dijkman R, Jebbink MF, Gaunt E, Rossen JW, Templeton KE, Kuijpers TW, et al. The dominance of human coronavirus OC43 and NL63 infections in infants. J Clin Virol. 2012;53(2):135–9.
Kalish H, Klumpp-Thomas C, Hunsberger S, Baus HA, Fay MP, Siripong N, et al. 2021, Mapping a Pandemic: SARS-CoV-2 Seropositivity in the United States. medRxiv. :https://doi.org/10.1101/2021.01.27.21250570.
Klumpp-Thomas C, Kalish H, Hicks J, Mehalko J, Drew M, Memoli MJ, et al. D614G spike variant does not Alter IgG, IgM, or IgA spike Seroassay performance. J Infect Dis. 2020.
Pallesen J, Wang N, Corbett KS, Wrapp D, Kirchdoerfer RN, Turner HL, et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci. 2017;114(35):E7348–E57.
Freeman B, Lester S, Mills L, Rasheed MAU, Moye S, Abiona O, et al. 2020, Validation of a SARS-CoV-2 spike protein ELISA for use in contact investigations and serosurveillance. bioRxiv.
Acknowledgements
The authors would like to thank Golan Ben-Oni, Rabbi Shua Brook, Dr. Adam Polinger, Dr. Avi Rosenberg, and the Jewish community of New York and New Jersey for their generous donation of blood samples use in this assay. We thank members of the FNLCR Protein Expression Laboratory (William Gillette, Simon Messing, and Vanessa Wall) for support in DNA production and protein purification.
Availability of Data and Materials
Raw data for the associated figures are available from the authors at reasonable request of the reader.
Funding
This research was supported in part by the Intramural Research Program of the NIH, including the National Institute of Biomedical Imaging and Bioengineering, the National Institute of Allergy and Infectious Diseases, and the National Center for Advancing Translational Sciences. This project has been funded in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. Disclaimer: The NIH, its officers, and employees do not recommend or endorse any company, product, or service.
Author information
Authors and Affiliations
Contributions
J.H., C.K.T., H.K., A.S., J.M., J.P.D, K.R.S., M.D., and K.S. performed experiments. M.J.M. provided control clinical samples. K.C., B.G., M.D.H., M.J.M., D.E., and K.S. provided guidance on experimental design and reagents. K.S. supervised the study.
Corresponding author
Ethics declarations
Ethical Approval and Consent Participate
All samples tested were fully de-identified and uncoded; this research fell under NIH IRB Exemption and did not require IRB approvals or clinical trial registration.
Consent for Publication
Not applicable.
Competing Interests
The authors declared that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(PDF 564 kb)
Rights and permissions
About this article
Cite this article
Hicks, J., Klumpp-Thomas, C., Kalish, H. et al. Serologic Cross-Reactivity of SARS-CoV-2 with Endemic and Seasonal Betacoronaviruses. J Clin Immunol 41, 906–913 (2021). https://doi.org/10.1007/s10875-021-00997-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-021-00997-6