Abstract
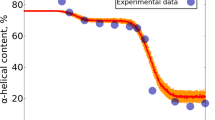
Glycoproteins are formed as the result of enzymatic glycosylation or chemical glycation in the body, and produced in vitro in industrial processes. The covalently attached carbohydrate molecule(s) confer new properties to the protein, including modified stability. In the present study, the structural stability of a glycoprotein form of myoglobin, bearing a glucose unit in the N-terminus, has been compared with its native form by the use of molecular dynamics simulation. Both structures were subjected to temperatures of 300 and 500 K in an aqueous environment for 10 ns. Changes in secondary structures and RMSD were then assessed. An overall higher stability was detected for glycomyoglobin, for which the most stable segments/residues were highlighted and compared with the native form. The simple addition of a covalently bound glucose is suggested to exert its stabilizing effect via increased contacts with surrounding water molecules, as well as a different pattern of interactions with neighbor residues.










Similar content being viewed by others
References
Capece, L., Boechi, L., Perissinotti, L.L., Arroyo-Manez, P., Bikiel, D.E., Smulevich, G., Marti, M.A., Estrin, D.A.: Small ligand-globin interactions: reviewing lessons derived from computer simulation. Biochim. Biophys. Acta 1834, 1722–1738 (2013)
Vinogradov, S.N., Hoogewijs, D., Bailly, X., Arredondo-Peter, R., Gough, J., Dewilde, S., Moens, L., Vanfleteren, J.R.: A phylogenomic profile of globins. BMC Evol. Biol. 6, 31 (2006)
Merlino, A., Vergara, A., Sica, F., Aschi, M., Amadei, A., Di Nola, A., Mazzarella, L.: Free-energy profile for CO binding to separated chains of human and Trematomus newnesi hemoglobin: insights from molecular dynamics simulations and perturbed matrix method. J. Phys. Chem. B 114, 7002–7008 (2010)
Ordway, G.A., Garry, D.J.: Myoglobin: an essential hemoprotein in striated muscle. J. Exp. Biol. 207, 3441–3446 (2004)
Chu, K., Vojtchovsky, J., McMahon, B.H., Sweet, R.M., Berendzen, J., Schlichting, I.: Structure of a ligand-binding intermediate in wild-type carbonmonoxy myoglobin. Nature 403, 921–923 (2000)
Ganz, T., Nemeth, E.: Regulation of iron acquisition and iron distribution in mammals. Biochim. Biophys. Acta 1763, 690–699 (2006)
Kendrew, J.C., Bodo, G., Dintzis, H.M., Parrish, R., Wyckoff, H., Phillips, D.: A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature 181, 662–666 (1958)
Stadler, A.M., Pellegrini, E., Johnson, M., Fitter, J., Zaccai, G.: Dynamics-stability relationships in apo- and holomyoglobin: a combined neutron scattering and molecular dynamics simulations study. Biophys. J. 102, 351–359 (2012)
Han, K.K., Martinage, A.: Post-translational chemical modification(s) of proteins. Int. J. Biochem. 24, 19–28 (1992)
Anguizola, J., Matsuda, R., Barnaby, O.S., Hoy, K.S., Wa, C., DeBolt, E., Koke, M., Hage, D.S.: Review: Glycation of human serum albumin. Clin. Chim. Acta 425, 64–76 (2013)
Nagai, R., Mori, T., Yamamoto, Y., Kaji, Y., Yonei, Y.: Significance of advanced glycation end products in aging-related disease. Anti-Aging. Med. 7, 112–119 (2010)
Han, M., Wang, X., Ding, H., Jin, M., Yu, L., Wang, J., Yu, X.: The role of N-glycosylation sites in the activity, stability, and expression of the recombinant elastase expressed by Pichia pastoris. Enzyme. Microb. Technol. 54, 32–37 (2014)
Wautier, J.-L., Guillausseau, P.-J.: Diabetes, advanced glycation endproducts and vascular disease. Vasc. Med. 3, 131–137 (1998)
Peppa, M., Uribarri, J., Vlassara, H.: Glucose, advanced glycation end products, and diabetes complications: what is new and what works. Clin. Diab. 21, 186–187 (2003)
O’Connor, S.E., Imperiali, B.: Modulation of protein structure and function by asparagine-linked glycosylation. Chem. Biol. 3, 803–812 (1996)
Vitek, M.P., Bhattacharya, K., Glendening, J.M., Stopa, E., Vlassara, H., Bucala, R., Manogue, K., Cerami, A.: Advanced glycation end products contribute to amyloidosis in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 91, 4766–4770 (1994)
Goldin, A., Beckman, J.A., Schmidt, A.M., Creager, M.A.: Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation 114, 597–605 (2006)
Stivala, A., Wybrow, M., Wirth, A., Whisstock, J.C., Stuckey, P.J.: Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics 27, 3315–3316 (2011)
Beck, D.A., Daggett, V.: Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods 34, 112–120 (2004)
Krishnan, A., Giuliani, A., Zbilut, J.P., Tomita, M.: Implications from a network-based topological analysis of ubiquitin unfolding simulations. PLoS ONE 3, e2149 (2008)
Wormald, M.R., Dwek, R.A.: Glycoproteins: glycan presentation and protein-fold stability. Structure 7, R155–160 (1999)
Jafari-Aghdam, J., Khajeh, K., Ranjbar, B., Nemat-Gorgani, M.: Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Biochim. Biophys. Acta 1750, 61–68 (2005)
Sola, R.J., Griebenow, K.: Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 98, 1223–1245 (2009)
Johansen, M.B., Kiemer, L., Brun, S.: Analysis and prediction of mammalian protein glycation. Glycobiology 16, 844–853 (2006)
Watkins, N.G., Thorpe, S.R., Baynes, J.W.: Glycation of amino groups in protein. J. Biol. Chem. 260, 10629–10636 (1985)
Sen, S., Bose, T., Roy, A., Chakraborti, A.S.: Effect of non-enzymatic glycation on esterase activities of hemoglobin and myoglobin. Mol. Cell. Biochem. 301, 251–257 (2007)
Iannuzzi, C., Maritato, R., Irace, G., Sirangelo, I.: Glycation accelerates fibrillization of the amyloidogenic W7FW14F apomyoglobin. PLoS ONE 8, e80768 (2013)
Roy, A., Sil, R., Chakraborti, A.S.: Non-enzymatic glycation induces structural modifications of myoglobin. Mol. Cell. Biochem. 338, 105–114 (2010)
Ansari, N.A., Dash, D.: Amadori glycated proteins: role in production of autoantibodies in diabetes mellitus and effect of inhibitors on non-enzymatic glycation. Aging Dis. 4, 50–6 (2013)
Cussimanio, B.L., Booth, A.A., Todd, P., Hudson, B.G., Khalifah, R.G.: Unusual susceptibility of heme proteins to damage by glucose during non-enzymatic glycation. Biophys. Chem. 105, 743–55 (2003)
Azami-Movahed, M., Shariatizi, S., Sabbaghian, M., Ghasemi, A., Ebrahim-Habibi, A., Nemat-Gorgani, M.: Heme binding site in apomyoglobin may be effectively targeted with small molecules to control aggregation. Int. J. Biochem. Cell Biol. 45, 299–307 (2013)
Banerjee, S., Chakraborti, A.S.: Structural alterations of hemoglobin and myoglobin by glyoxal: a comparative study. Int. J. Biol. Macromol. 66, 311–8 (2014)
Acknowledgments
This study was supported by the Endocrinology and Metabolism Research Institute and performed in its Modeling and Simulation Lab.
Conflict of interest
The authors declare that they have no conflicts of interest to disclose.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Snapshots of native and glycated form of human myoglobin simulations extracted at first and last time points at 500 K. Ribbons are α-helices. (GIF 54 kb)
Fig. S2
Three-dimensional representation of residues located at 4.5 Å of Gly1 in (a) Mb and (b) GMb over time in ten sampled structures. (GIF 122 kb)
(GIF 125 kb)
Rights and permissions
About this article
Cite this article
Alizadeh-Rahrovi, J., Shayesteh, A. & Ebrahim-Habibi, A. Structural stability of myoglobin and glycomyoglobin: a comparative molecular dynamics simulation study. J Biol Phys 41, 349–366 (2015). https://doi.org/10.1007/s10867-015-9383-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10867-015-9383-2