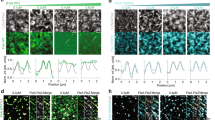
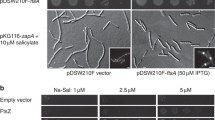
Abstract
Fission of many prokaryotes as well as some eukaryotic organelles depends on the self-assembly of the FtsZ protein into a membrane-associated ring structure early in the division process. Different components of the machinery are then sequentially recruited. Although the assembly order has been established, the molecular interactions and the understanding of the force-generating mechanism of this dividing machinery have remained elusive. It is desirable to develop simple reconstituted systems that attempt to reproduce, at least partially, some of the stages of the process. High-resolution studies of Escherichia coli FtsZ filaments’ structure and dynamics on mica have allowed the identification of relevant interactions between filaments that suggest a mechanism by which the polymers could generate force on the membrane. Reconstituting the membrane-anchoring protein ZipA on E. coli lipid membrane on surfaces is now providing information on how the membrane attachment regulates FtsZ polymer dynamics and indicates the important role played by the lipid composition of the membrane.





Similar content being viewed by others
References
Harry, E., Monahan, L., Thompson, L.: Bacterial cell division: the mechanism and its precision. Int. Rev. Cytol. 253, 27–93 (2006). doi:10.1016/S0074-7696(06)53002-5
Vicente, M., Rico, A.I.: The order of the ring: assembly of Escherichia coli cell division components. Mol. Microbiol. 61(1), 5–8 (2006)
Surrey, T., Nédélec, F., Leibler, S., Karsenti, E.: Physical properties determining self-organization of motors and microtubules. Science 292(5519), 1167–1171 (2001). doi:10.1126/science.1059758
Karsenti, E., Nédélec, F., Surrey, T.: Modelling microtubule patterns. Nat. Cell Biol. 8(11), 1204–1211 (2006). doi:10.1038/ncb1498
Leduc, C., Campas, O., Zeldovich, K.B., Roux, A., Jolimaitre, P., Bourel-Bonnet, L., Goud, B., Joanny, J.F., Bassereau, P., Prost, J.: Cooperative extraction of membrane nanotubes by molecular motors. Proc. Natl. Acad. Sci. U. S. A. 101(49), 17096–17101 (2004). doi:10.1073/pnas.0406598101
Footer, M.J., Kerssemakers, J.W.J., Theriot, J.A., Dogterom, M.: Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl. Acad. Sci. U. S. A. 104(7), 2181–2186 (2007). doi:10.1073/pnas.0607052104
Lagomarsino, M.C., Tanase, C., Vos, J.W., Emons, A.M.C., Mulder, B.M., Dogterom, M.: Microtubule organization in three-dimensional confined geometries: evaluating the role of elasticity through a combined in vitro and modeling approach. Biophys. J. 92(3), 1046–1057 (2007). doi:10.1529/biophysj.105.076893
Brunner, C., Wahnes, C., Vogel, V.: Cargo pick-up from engineered loading stations by kinesin driven molecular shuttles. Lab Chip 7, 1263–1271 (2007). doi:10.1039/b707301a
Clemmens, J., Hess, H., Doot, R., Matzke, C.M., Bachand, G.D., Vogel, V.: Motor-protein “roundabouts”: microtubules moving on kinesin-coated tracks through engineered networks. Lab Chip 4, 83–86 (2004)
Shih, Y.-L., Rothfield, L.: The bacterial cytoskeleton. Microbiol. Mol. Biol. Rev. 70(3), 729–754 (2006). doi:10.1128/MMBR.00017-06
Dai, K., Lutkenhaus, J.: FtsZ is an essential cell division gene in Escherichia coli. J. Bacteriol. 173(11), 3500–3506 (1991)
Margolin, W.: FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862–871 (2005). doi:10.1038/nrm1745
Dajkovic, A., Lutkenhaus, J.: Z ring as executor of bacterial cell division. J. Mol. Microbiol. Biotechnol. 11, 140–151 (2006). doi:10.1159/000094050
Romberg, L., Levin, P.A.: Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57, 125–154 (2003). doi:10.1146/annurev.micro.57.012903.074300
Stricker, J., Maddox, P., Salmon, E.D., Erickson, H.P.: Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. U. S. A. 99, 3171–3175 (2002). doi:10.1073/pnas.052595099
Hale, C.A., de Boer, P.A.J.: Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997). doi:10.1016/S0092-8674(00)81838-3
Rivas, G., López, A., Mingorance, J., Ferrandiz, M.J., Zorrilla, S., Minton, A., Vicente, M., Andreu, J.M.: Magnesium-induced linear self-association of the FtsZ bacterial cell division protein monomer—the primary steps for the FtsZ assembly. J. Biol. Chem. 275, 11740–11749 (2000). doi:10.1074/jbc.275.16.11740
RayChaudhuri, D.: ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18, 2372–2383 (1999). doi:10.1093/emboj/18.9.2372
Moreno-Herrero, F., de Pablo, P.J., Fernández-Sánchez, R., Colchero, J., Gómez-Herrero, J., Baró, A.M.: Scanning force microscopy jumping and tapping modes in liquids. Appl. Phys. Lett. 81(14), 2620–2622 (2002). doi:10.1063/1.1509856
Levy, D.C.M., Rigaud, J.L.: Two-dimensional crystallization of membrane proteins: the lipid layer strategy. FEBS Lett. 504, 187–193 (2001). doi:10.1016/S0014-5793(01)02748-X
Mingorance, J., Tadros, M., Vicente, M., González, J.M., Rivas, G., Vélez, M.: Visualization of single Escherichia coli FtsZ filament dynamics with atomic force microscopy. J. Biol. Chem. 280, 20909–20914 (2005). doi:10.1074/jbc.M503059200
González, J.M., Vélez, M., Jiménez, M., Alfonso, C., Schuck, P., Mingorance, J., Vicente, M., Minton, A.P., Rivas, G.: The cooperative behavior of E. coli cell division protein FtsZ assembly involves the preferential cyclization of long single-stranded fibrils. Proc. Natl. Acad. Sci. U. S. A. 102, 1895–1900 (2005). doi:10.1073/pnas.0409517102
Carlier, M., Didry, D., Melki, R., Chabre, M., Pantaloni, D.: Stabilization of microtubules by inorganic phosphate and its structural analogues, the fluoride complexes of aluminium and beryllium. Biochemistry 28, 3628 (1989). doi:10.1021/bi00434a073
Bigay, J.D.P., Pfister, C., Chabre, M.: Fluoride complexes of aluminium or beryllium act on G-proteins as reversibly bound analogues of the gamma phosphate of GTP. EMBO J. 6, 2907–2913 (1987)
RayChaudhuri, D., Park, J.T.: A point mutation converts Escherichia coli FtsZ septation GTPase to an ATPase. J. Biol. Chem. 269, 22941–22944 (1994)
Hörger, I., Velasco, E., Mingorance, J., Rivas, G., Vélez, M., Tarazona, P.: Langevin computer simulations of FtsZ filaments and the force generating mechanism during cell division. Phys. Rev., E 77, 011902 (2008)
Pichoff, S., Lutkenhaus, J.: Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55(6), 1722–1734 (2005). doi:10.1111/j.1365-2958.2005.04522.x
Ohashi, T., Hale, C.A., de Boer, P.A.J., Erickson, H.P.: Structural evidence that the P/Q domain of ZipA is an unstructured, flexible tether between the membrane and the C-terminal FtsZ-binding domain. J. Bacteriol. 184(15), 4313–4315 (2002). doi:10.1128/JB.184.15.4313-4315.2002
Hale, C.A., Rhee, A.C., de Boer, P.A.J.: ZipA-Induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182(18), 5153–5166 (2000). doi:10.1128/JB.182.18.5153-5166.2000
Erickson, H., Taylor, D., Taylor, K.A., Bramhill, D.: Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl. Acad. Sci. U. S. A. 93, 519–523 (1996). doi:10.1073/pnas.93.1.519
Díaz, J.F., Kralicek, A., Mingorance, J., Palacios, J.M., Vicente, M., Andreu, J.M.: Activation of cell division protein FtsZ. J. Biol. Chem. 276(20), 17307–17315 (2001). doi:10.1074/jbc.M010920200
Hörger, I., Velasco, E., Rivas, G., Vélez, M., Tarazona, P.: FtsZ bacterial cytoskeletal polymers on curved surfaces: the importance of lateral interactions. Biophys. J. 94, L81–L83 (2008). doi:10.1529/biophysj.107.128363
Lu, C., Stricker, J., Erickson, H.P.: Site-specific mutations of FtsZ—effects on GTPase and in vitro assembly. BMC Microbiol. 1(7), 1471–1482 (2001)
Esue, O., Tseng, Y., Wirtz, D.: The rapid onset of elasticity during the assembly of the bacterial cell-division protein FtsZ. Biochem. Biophys. Res. Commun. 333, 508–516 (2005). doi:10.1016/j.bbrc.2005.05.152
Andrews, S.S., Arkin, A.P.: A mechanical explanation for cytoskeletal rings and helices in bacteria. Biophys. J. 93, 1872–1884 (2007). doi:10.1529/biophysj.106.102343
Erickson, H.P.: The FtsZ protofilament and attachment of ZipA—structural constraints on the FtsZ power stroke. Curr. Opin. Cell Biol. 13, 55–60 (2001). doi:10.1016/S0955-0674(00)00174-5
Acknowledgements
This work was financed by the Ministerio de Educación y Ciencia (MEC) under grants BFU2005-04087-C02, FIS2004-05035-C03-02, BCM2002-04617-C02-02 and the Comunidad Autónoma de Madrid (CAM) under NANOBIO-M (http://www.nanobiom.org), grant S-0505/MAT0283 and grant S-0505/ESP-0299.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Navajas, P.L., Rivas, G., Mingorance, J. et al. In Vitro Reconstitution of the Initial Stages of the Bacterial Cell Division Machinery. J Biol Phys 34, 237–247 (2008). https://doi.org/10.1007/s10867-008-9118-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10867-008-9118-8