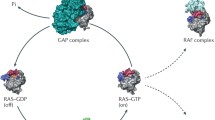
Abstract
Mutations in RAS oncogenes occur in ~ 30% of human cancers, with KRAS being the most frequently altered isoform. RAS proteins comprise a conserved GTPase domain and a C-terminal lipid-modified tail that is unique to each isoform. The GTPase domain is a ‘switch’ that regulates multiple signaling cascades that drive cell growth and proliferation when activated by binding GTP, and the signal is terminated by GTP hydrolysis. Oncogenic RAS mutations disrupt the GTPase cycle, leading to accumulation of the activated GTP-bound state and promoting proliferation. RAS is a key target in oncology, however it lacks classic druggable pockets and has been extremely challenging to target. RAS signaling has thus been targeted indirectly, by harnessing key downstream effectors as well as upstream regulators, or disrupting the proper membrane localization required for signaling, by inhibiting either lipid modification or ‘carrier’ proteins. As a small (20 kDa) protein with multiple conformers in dynamic equilibrium, RAS is an excellent candidate for NMR-driven characterization and screening for direct inhibitors. Several molecules have been discovered that bind RAS and stabilize shallow pockets through conformational selection, and recent compounds have achieved substantial improvements in affinity. NMR-derived insight into targeting the RAS-membrane interface has revealed a new strategy to enhance the potency of small molecules, while another approach has been development of peptidyl inhibitors that bind through large interfaces rather than deep pockets. Remarkable progress has been made with mutation-specific covalent inhibitors that target the thiol of a G12C mutant, and these are now in clinical trials. Here we review the history of RAS inhibitor development and highlight the utility of NMR and integrated biophysical approaches in RAS drug discovery.





Similar content being viewed by others
References
Agazie YM, Hayman MJ (2003) Molecular mechanism for a role of SHP2 in epidermal growth factor receptor signaling. Mol Cell Biol 23:7875–7886
Ahearn IM, Haigis K, Bar-Sagi D, Philips MR (2011) Regulating the regulator: post-translational modification of RAS. Nat Rev Mol Cell Biol 13:39–51
Ahmed TA et al (2019) SHP2 Drives Adaptive Resistance to ERK Signaling Inhibition in Molecularly Defined Subsets of ERK-Dependent Tumors. Cell Rep 26:65–78
Alvarez-Moya B, Barcelo C, Tebar F, Jaumot M, Agell N (2011) CaM interaction and Ser181 phosphorylation as new K-Ras signaling modulators. Small GTPases 2:99–103
Antic I, Biancucci M, Zhu Y, Gius DR, Satchell KJF (2015) Site-specific processing of Ras and Rap1 Switch I by a MARTX toxin effector domain. Nat Commun 6:7396
Baines AT, Xu D, Der CJ (2011) Inhibition of Ras for cancer treatment: the search continues. Future Med Chem 3:1787–1808
Barbacid M (1987) ras genes. Annu Rev Biochem 56:779–827
Barr AJ (2010) Protein tyrosine phosphatases as drug targets: strategies and challenges of inhibitor development. Future Med Chem 2:1563–1576
Basso AD, Kirschmeier P, Bishop WR (2006) Lipid posttranslational modifications. Farnesyl transferase inhibitors. J Lipid Res 47:15–31
Bennett AM, Tang TL, Sugimoto S, Walsh CT, Neel BG (1994) Protein-tyrosine-phosphatase SHPTP2 couples platelet-derived growth factor receptor beta to Ras. Proc Natl Acad Sci USA 91:7335–7339
Bentires-Alj M et al (2004) Activating mutations of the noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res 64:8816–8820
Berndt N, Hamilton AD, Sebti SM (2011) Targeting protein prenylation for cancer therapy. Nat Rev Cancer 11:775–791
Bery N et al (2018) BRET-based RAS biosensors that show a novel small molecule is an inhibitor of RAS-effector protein-protein interactions. Elife 7:e37122
Bery N et al (2019) KRAS-specific inhibition using a DARPin binding to a site in the allosteric lobe. Nat Commun 10:2607
Bery N, Miller A, Rabbitts T (2020) A potent KRAS macromolecule degrader specifically targeting tumours with mutant KRAS. Nat Commun 11:3233
Bhagatji P, Leventis R, Rich R, Lin CJ, Silvius JR (2010) Multiple cellular proteins modulate the dynamics of K-ras association with the plasma membrane. Biophys J 99:3327–3335
Binz HK, Stumpp MT, Forrer P, Amstutz P, Pluckthun A (2003) Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J Mol Biol 332:489–503
Bivona TG et al (2006) PKC regulates a farnesyl-electrostatic switch on K-Ras that promotes its association with Bcl-XL on mitochondria and induces apoptosis. Mol Cell 21:481–493
Bollag G et al (2012) Vemurafenib: the first drug approved for BRAF-mutant cancer. Nat Rev Drug Discov 11:873–886
Bos JL (1989) ras oncogenes in human cancer: a review. Cancer Res 49:4682–4689
Brown JS, Banerji U (2017) Maximising the potential of AKT inhibitors as anti-cancer treatments. Pharmacol Ther 172:101–115
Brunsveld L et al (2006) Lipidated ras and rab peptides and proteins–synthesis, structure, and function. Angew Chem Int Ed Engl 45:6622–6646
Bunda S et al (2014) Src promotes GTPase activity of Ras via tyrosine 32 phosphorylation. Proc Natl Acad Sci USA 111:E3785–E3794
Bunda S et al (2015) Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat Commun 6:8859
Campbell-Burk SL, Domaille PJ, Starovasnik MA, Boucher W, Laue ED (1992) Sequential assignment of the backbone nuclei (1H, 15N and 13C) of c-H-ras p21 (1–166). GDP using a novel 4D NMR strategy. J Biomol NMR 2:639–646
Canon J et al (2019) The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575:217–223
Caunt CJ, Sale MJ, Smith PD, Cook SJ (2015) MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 15:577–592
Ceska T, Chung CW, Cooke R, Phillips C, Williams PA (2019) Cryo-EM in drug discovery. Biochem Soc Trans 47:281–293
Chandra A et al (2011) The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol 14:148–158
Chen X, Makarewicz JM, Knauf JA, Johnson LK, Fagin JA (2014) Transformation by Hras(G12V) is consistently associated with mutant allele copy gains and is reversed by farnesyl transferase inhibition. Oncogene 33:5442–5449
Chen YN et al (2016) Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature 535:148–152
Chen D et al (2019) Fragment-based drug discovery of triazole inhibitors to block PDEdelta-RAS protein-protein interaction. Eur J Med Chem 163:597–609
Cho KJ et al (2016) AMPK and endothelial nitric oxide synthase signaling regulates K-Ras plasma membrane interactions via cyclic GMP-dependent protein kinase 2. Mol Cell Biol 36:3086–3099
Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ (2014) Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov 13:828–851
Cox AD, Der CJ, Philips MR (2015) Targeting RAS membrane association: back to the future for anti-RAS drug discovery? Clin Cancer Res 21:1819–1827
Cruz-Migoni A et al (2019) Structure-based development of new RAS-effector inhibitors from a combination of active and inactive RAS-binding compounds. Proc Natl Acad Sci USA 116:2545–2550
Dalvit C et al (2000) Identification of compounds with binding affinity to proteins via magnetization transfer from bulk water. J Biomol NMR 18:65–68
Dance M, Montagner A, Salles JP, Yart A, Raynal P (2008) The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell Signal 20:453–459
Dardaei L et al (2018) SHP2 inhibition restores sensitivity in ALK-rearranged non-small-cell lung cancer resistant to ALK inhibitors. Nat Med 24:512–517
De Roock W et al (2010) Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA 304:1812–1820
Denisov IG, Grinkova YV, Lazarides AA, Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J Am Chem Soc 126:3477–3487
Dharmaiah S et al (2016) Structural basis of recognition of farnesylated and methylated KRAS4b by PDEdelta. Proc Natl Acad Sci USA 113:E6766–E6775
Donohue E et al (2019) Second harmonic generation detection of Ras conformational changes and discovery of a small molecule binder. Proc Natl Acad Sci USA 116:17290–17297
Durrant DE, Morrison DK (2018) Targeting the Raf kinases in human cancer: the Raf dimer dilemma. Br J Cancer 118:3–8
Eglen RM et al (2008) The use of AlphaScreen technology in HTS: current status. Curr Chem Genomics 1:2–10
Fang Z et al (2018) Inhibition of K-RAS4B by a unique mechanism of action: stabilizing membrane-dependent occlusion of the effector-binding site. Cell Chem Biol 25:1327–1336
Fang Z et al (2020) Multivalent assembly of KRAS with the RAS-binding and cysteine-rich domains of CRAF on the membrane. Proc Natl Acad Sci USA 117:12101–12108
Feng H et al (2019) K-Ras(G12D) has a potential allosteric small molecule binding site. Biochemistry 58:2542–2554
Fivaz M, Meyer T (2005) Reversible intracellular translocation of KRas but not HRas in hippocampal neurons regulated by Ca2+/calmodulin. J Cell Biol 170:429–441
Ford B, Skowronek K, Boykevisch S, Bar-Sagi D, Nassar N (2005) Structure of the G60A mutant of Ras: implications for the dominant negative effect. J Biol Chem 280:25697–25705
Gai SA, Wittrup KD (2007) Yeast surface display for protein engineering and characterization. Curr Opin Struct Biol 17:467–473
Gajate P et al (2012) Influence of KRAS p.G13D mutation in patients with metastatic colorectal cancer treated with cetuximab. Clin Colorectal Cancer 11:291–296
Gehringer M, Laufer SA (2019) Emerging and re-emerging warheads for targeted covalent inhibitors: applications in medicinal chemistry and chemical biology. J Med Chem 62:5673–5724
Geyer M et al (1996) Conformational transitions in p21ras and in its complexes with the effector protein Raf-RBD and the GTPase activating protein GAP. Biochemistry 35:10308–10320
Ghosh AK, Samanta I, Mondal A, Liu WR (2019) Covalent inhibition in drug discovery. ChemMedChem 14:889–906
Go ML et al (2010) Amino derivatives of indole as potent inhibitors of isoprenylcysteine carboxyl methyltransferase. J Med Chem 53:6838–6850
Grant BMM et al (2020) Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety. Sci Signal 13(625):eaaz0344
Guillard S et al (2017) Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange. Nat Commun 8:16111
Gupta AK et al (2019) Multi-target, ensemble-based virtual screening yields novel allosteric KRAS inhibitors at high success rate. Chem Biol Drug Des 94:1441–1456
Ha JM et al (1989) Conformation of guanosine 5'-diphosphate as bound to a human c-Ha-ras mutant protein: a nuclear Overhauser effect study. Biochemistry 28:8411–8416
Hallin J et al (2020) The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 10:54–71
Hanafusa H, Torii S, Yasunaga T, Nishida E (2002) Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol 4:850–858
Hancock JF, Magee AI, Childs JE, Marshall CJ (1989) All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57:1167–1177
Hancock JF, Paterson H, Marshall CJ (1990) A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63:133–139
Hillig RC et al (2019) Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction. Proc Natl Acad Sci USA 116:2551–2560
Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE (1998) Crystal structure of the tyrosine phosphatase SHP-2. Cell 92:441–450
Holderfield M (2018) Efforts to develop KRAS inhibitors. Cold Spring Harb Perspect Med 8:a031864
Hunter JC et al (2015) Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res 13:1325–1335
Ismail SA et al (2011) Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol 7:942–949
Ito Y et al (1997) Regional polysterism in the GTP-bound form of the human c-Ha-Ras protein. Biochemistry 36:9109–9119
Jackson JH, Li JW, Buss JE, Der CJ, Cochrane CG (1994) Polylysine domain of K-ras 4B protein is crucial for malignant transformation. Proc Natl Acad Sci USA 91:12730–12734
Jansen JM et al (2017) Inhibition of prenylated KRAS in a lipid environment. PLoS ONE 12:e0174706
John J et al (1990) Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry 29:6058–6065
Jura N, Scotto-Lavino E, Sobczyk A, Bar-Sagi D (2006) Differential modification of Ras proteins by ubiquitination. Mol Cell 21:679–687
Kalbitzer HR, Spoerner M (2013) State 1(T) inhibitors of activated Ras. Enzymes 33(Pt A):69–94
Kalbitzer HR, Spoerner M, Ganser P, Hozsa C, Kremer W (2009) Fundamental link between folding states and functional states of proteins. J Am Chem Soc 131:16714–16719
Kano Y et al (2019) Tyrosyl phosphorylation of KRAS stalls GTPase cycle via alteration of switch I and II conformation. Nat Commun 10:224
Kauke MJ et al (2017) An engineered protein antagonist of K-Ras/B-Raf interaction. Sci Rep 7:5831
Kemp DS, Chien SW (1967) A new peptide coupling reagent. J Am Chem Soc 89:2743–2745
Kessler D et al (2019) Drugging an undruggable pocket on KRAS. Proc Natl Acad Sci USA 116:15823–15829
Kessler D et al (2020) Reply to Tran et al.: Dimeric KRAS protein-protein interaction stabilizers. Proc Natl Acad Sci USA 117:3365–3367
Khan I, Spencer-Smith R, O'Bryan JP (2019) Targeting the alpha4-alpha5 dimerization interface of K-RAS inhibits tumor formation in vivo. Oncogene 38:2984–2993
Khan I, Rhett JM, O'Bryan JP (2020) Therapeutic targeting of RAS: New hope for drugging the "undruggable". Biochim Biophys Acta Mol Cell Res 1867:118570
Kidger AM, Sipthorp J, Cook SJ (2018) ERK1/2 inhibitors: New weapons to inhibit the RAS-regulated RAF-MEK1/2-ERK1/2 pathway. Pharmacol Ther 187:45–60
Kondo Y et al (2019) Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases. Science 366:109–115
Laheru D et al (2012) Integrated preclinical and clinical development of S-trans, trans-Farnesylthiosalicylic Acid (FTS, Salirasib) in pancreatic cancer. Invest New Drugs 30:2391–2399
Lander HM et al (1997) A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J Biol Chem 272:4323–4326
Lavoie H, Therrien M (2015) Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 16:281–298
Lee KY et al (2020) Two distinct structures of membrane-associated homodimers of GTP- and GDP-bound KRAS4B revealed by paramagnetic relaxation enhancement. Angew Chem Int Ed Engl 59:11037–11045
Leshchiner ES et al (2015) Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci USA 112:1761–1766
Leventis R, Silvius JR (1998) Lipid-binding characteristics of the polybasic carboxy-terminal sequence of K-ras4B. Biochemistry 37:7640–7648
Li Z, Buck M (2020) Computational design of myristoylated cell-penetrating peptides targeting oncogenic K-Ras.G12D at the effector-binding membrane interface. J Chem Inf Model 60:306–315
Li S, Jang H, Zhang J, Nussinov R (2018a) Raf-1 cysteine-rich domain increases the affinity of K-Ras/Raf at the membrane, promoting MAPK signaling. Structure 26:513–525
Li ZL, Prakash P, Buck M (2018b) A "Tug of War" maintains a dynamic protein-membrane complex: molecular dynamics simulations of C-Raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent Sci 4:298–305
Lim SM et al (2014) Therapeutic targeting of oncogenic K-Ras by a covalent catalytic site inhibitor. Angew Chem Int Ed Engl 53:199–204
Lobell RB et al (2002) Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol Cancer Ther 1:747–758
Lopez-Alcala C et al (2008) Identification of essential interacting elements in K-Ras/calmodulin binding and its role in K-Ras localization. J Biol Chem 283:10621–10631
Lu S, Jang H, Nussinov R, Zhang J (2016) The structural basis of oncogenic mutations G12, G13 and Q61 in small GTPase K-Ras4B. Sci Rep 6:21949
Mainardi S et al (2018) SHP2 is required for growth of KRAS-mutant non-small-cell lung cancer in vivo. Nat Med 24:961–967
Markham A (2019) Alpelisib: first global approval. Drugs 79:1249–1253
Martin-Gago P et al (2017a) Covalent protein labeling at glutamic acids. Cell Chem Biol 24:589–597
Martin-Gago P et al (2017b) A PDE6delta-KRas inhibitor chemotype with up to seven H-bonds and picomolar affinity that prevents efficient inhibitor release by Arl2. Angew Chem Int Ed Engl 56:2423–2428
Matsumoto S et al (2018) Molecular basis for allosteric inhibition of GTP-bound H-Ras protein by a small-molecule compound carrying a naphthalene ring. Biochemistry 57:5350–5358
Maurer T et al (2012) Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci USA 109:5299–5304
Mayer M, Meyer B (2001) Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J Am Chem Soc 123:6108–6117
Mazhab-Jafari MT et al (2015) Oncogenic and RASopathy-associated K-RAS mutations relieve membrane-dependent occlusion of the effector-binding site. Proc Natl Acad Sci USA 112:6625–6630
McCarthy MJ et al (2019) Discovery of high-affinity noncovalent allosteric KRAS inhibitors that disrupt effector binding. ACS Omega 4:2921–2930
McGee JH et al (2018) Exceptionally high-affinity Ras binders that remodel its effector domain. J Biol Chem 293:3265–3280
McLean MA, Stephen AG, Sligar SG (2019) PIP2 influences the conformational dynamics of membrane-bound KRAS4b. Biochemistry 58:3537–3545
Mendola CE, Backer JM (1990) Lovastatin blocks N-ras oncogene-induced neuronal differentiation. Cell Growth Differ 1:499–502
Millburn MV et al (1990) Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science 247:939–945
Montagner A et al (2005) A novel role for Gab1 and SHP2 in epidermal growth factor-induced Ras activation. J Biol Chem 280:5350–5360
Nagasaka M et al (2020) KRAS G12C Game of Thrones, which direct KRAS inhibitor will claim the iron throne? Cancer Treat Rev 84:101974
Nan X et al (2015) Ras-GTP dimers activate the Mitogen-Activated Protein Kinase (MAPK) pathway. Proc Natl Acad Sci USA 112:7996–8001
Nichols RJ et al (2018) RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 20:1064–1073
Noonan T, Brown N, Dudycz L, Wright G (1991) Interaction of GTP derivatives with cellular and oncogenic ras-p21 proteins. J Med Chem 34:1302–1307
Novotny CJ, Hamilton GL, McCormick F, Shokat KM (2017) Farnesyltransferase-mediated delivery of a covalent inhibitor overcomes alternative prenylation to mislocalize K-Ras. ACS Chem Biol 12:1956–1962
Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM (2013) K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature 503:548–551
Papke B et al (2016) Identification of pyrazolopyridazinones as PDEdelta inhibitors. Nat Commun 7:11360
Park E et al (2019) Architecture of autoinhibited and active BRAF-MEK1-14-3-3 complexes. Nature 575:545–550
Patgiri A, Jochim AL, Arora PS (2008) A hydrogen bond surrogate approach for stabilization of short peptide sequences in alpha-helical conformation. Acc Chem Res 41:1289–1300
Patgiri A, Yadav KK, Arora PS, Bar-Sagi D (2011) An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol 7:585–587
Patricelli MP et al (2016) Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 6:316–329
Prior IA, Lewis PD, Mattos C (2012) A comprehensive survey of Ras mutations in cancer. Cancer Res 72:2457–2467
Prior IA, Hood FE, Hartley JL (2020) The frequency of Ras mutations in cancer. Cancer Res
Quambusch L et al (2019) Covalent-allosteric inhibitors to achieve Akt isoform-selectivity. Angew Chem Int Ed Engl 58:18823–18829
Quatela SE, Sung PJ, Ahearn IM, Bivona TG, Philips MR (2008) Analysis of K-Ras phosphorylation, translocation, and induction of apoptosis. Methods Enzymol 439:87–102
Quevedo CE et al (2018) Small molecule inhibitors of RAS-effector protein interactions derived using an intracellular antibody fragment. Nat Commun 9:3169
Raponi M, Winkler H, Dracopoli NC (2008) KRAS mutations predict response to EGFR inhibitors. Curr Opin Pharmacol 8:413–418
Renaud JP et al (2018) Cryo-EM in drug discovery: achievements, limitations and prospects. Nat Rev Drug Discov 17:471–492
Riely GJ et al (2011) A phase II trial of Salirasib in patients with lung adenocarcinomas with KRAS mutations. J Thorac Oncol 6:1435–1437
Rocks O et al (2005) An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746–1752
Rocks O, Peyker A, Bastiaens PI (2006) Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Curr Opin Cell Biol 18:351–357
Rocks O et al (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141:458–471
Rosnizeck IC et al (2010) Stabilizing a weak binding state for effectors in the human ras protein by cyclen complexes. Angew Chem Int Ed Engl 49:3830–3833
Rosnizeck IC et al (2012) Metal-bis(2-picolyl)amine complexes as state 1(T) inhibitors of activated Ras protein. Angew Chem Int Ed Engl 51:10647–10651
Ruess DA et al (2018) Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat Med 24:954–960
Ryan MB, Der CJ, Wang-Gillam A, Cox AD (2015) Targeting RAS-mutant cancers: is ERK the key? Trends Cancer 1:183–198
Saito N, Mine N, Kufe DW, Von Hoff DD, Kawabe T (2017) CBP501 inhibits EGF-dependent cell migration, invasion and epithelial-to-mesenchymal transition of non-small cell lung cancer cells by blocking KRas to calmodulin binding. Oncotarget 8:74006–74018
Saur M et al (2019) Fragment-based drug discovery using cryo-EM. Drug Discov Today 61(3):543–560
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168:960–976
Schapira M, Calabrese MF, Bullock AN, Crews CM (2019) Targeted protein degradation: expanding the toolbox. Nat Rev Drug Discov 18:949–963
Schmick M et al (2014) KRas localizes to the plasma membrane by spatial cycles of solubilization, trapping and vesicular transport. Cell 157:459–471
Shima F et al (2010) Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem 285:22696–22705
Shima F et al (2013) In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras-effector interaction. Proc Natl Acad Sci USA 110:8182–8187
Shima F et al (2015) Current status of the development of Ras inhibitors. J Biochem 158:91–99
Shuker SB, Hajduk PJ, Meadows RP, Fesik SW (1996) Discovering high-affinity ligands for proteins: SAR by NMR. Science 274:1531–1534
Siddiqui FA et al (2020) PDE6D inhibitors with a new design principle selectively block K-Ras activity. ACS Omega 5:832–842
Silvius JR, l'Heureux F (1994) Fluorimetric evaluation of the affinities of isoprenylated peptides for lipid bilayers. Biochemistry 33:3014–3022
Silvius JR, Bhagatji P, Leventis R, Terrone D (2006) K-ras4B and prenylated proteins lacking "second signals" associate dynamically with cellular membranes. Mol Biol Cell 17:192–202
Simanshu DK, Nissley DV, McCormick F (2017) RAS Proteins and Their Regulators in Human Disease. Cell 170:17–33
Singh J, Petter RC, Baillie TA, Whitty A (2011) The resurgence of covalent drugs. Nat Rev Drug Discov 10:307–317
Smith GP (1985) Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 228:1315–1317
Smith MJ, Ikura M (2014) Integrated RAS signaling defined by parallel NMR detection of effectors and regulators. Nat Chem Biol 10:223–230
Smith MJ, Neel BG, Ikura M (2013) NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc Natl Acad Sci USA 110:4574–4579
Spencer-Smith R et al (2017) Inhibition of RAS function through targeting an allosteric regulatory site. Nat Chem Biol 13:62–68
Sperlich B, Kapoor S, Waldmann H, Winter R, Weise K (2016) Regulation of K-Ras4B Membrane Binding by Calmodulin. Biophys J 111:113–122
Spiegel J, Cromm PM, Zimmermann G, Grossmann TN, Waldmann H (2014) Small-molecule modulation of Ras signaling. Nat Chem Biol 10:613–622
Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A (2001) Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA 98:4944–4949
Spoerner M, Wittinghofer A, Kalbitzer HR (2004) Perturbation of the conformational equilibria in Ras by selective mutations as studied by 31P NMR spectroscopy. FEBS Lett 578:305–310
Spoerner M, Graf T, Konig B, Kalbitzer HR (2005) A novel mechanism for the modulation of the Ras-effector interaction by small molecules. Biochem Biophys Res Commun 334:709–713
Stephen AG, Esposito D, Bagni RK, McCormick F (2014) Dragging ras back in the ring. Cancer Cell 25:272–281
Sun Q et al (2012) Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl 51:6140–6143
Sun Q et al (2014) A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand. J Biomol NMR 60:11–14
Sung PJ et al (2013) Phosphorylated K-Ras limits cell survival by blocking Bcl-xL sensitization of inositol trisphosphate receptors. Proc Natl Acad Sci USA 110:20593–20598
Tejpar S et al (2012) Association of KRAS G13D tumor mutations with outcome in patients with metastatic colorectal cancer treated with first-line chemotherapy with or without cetuximab. J Clin Oncol 30:3570–3577
Thorpe LM, Yuzugullu H, Zhao JJ (2015) PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer 15:7–24
Tran TH et al (2020) The small molecule BI-2852 induces a nonfunctional dimer of KRAS. Proc Natl Acad Sci USA 117:3363–3364
Traut TW (1994) Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 140:1–22
Travers T et al (2018) Molecular recognition of RAS/RAF complex at the membrane: Role of RAF cysteine-rich domain. Sci Rep 8:8461
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754
Vidimar V et al (2020) An engineered chimeric toxin that cleaves activated mutant and wild-type RAS inhibits tumor growth. Proc Natl Acad Sci USA
Vigil D, Cherfils J, Rossman KL, Der CJ (2010) Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer 10:842–857
Villalonga P et al (2001) Calmodulin binds to K-Ras, but not to H- or N-Ras, and modulates its downstream signaling. Mol Cell Biol 21:7345–7354
Wahlstrom AM et al (2008) Inactivating Icmt ameliorates K-RAS-induced myeloproliferative disease. Blood 112:1357–1365
Wang MT et al (2015) K-Ras promotes tumorigenicity through suppression of non-canonical Wnt signaling. Cell 163:1237–1251
Welsch ME et al (2017) Multivalent small-molecule Pan-RAS inhibitors. Cell 168:878–889
Whyte DB et al (1997) K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem 272:14459–14464
Willumsen BM, Christensen A, Hubbert NL, Papageorge AG, Lowy DR (1984) The p21 ras C-terminus is required for transformation and membrane association. Nature 310:583–586
Winter-Vann AM, Casey PJ (2005) Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer 5:405–412
Wolfson E, Schmukler E, Schokoroy ST, Kloog Y, Pinkas-Kramarski R (2015) Enhancing FTS (Salirasib) efficiency via combinatorial treatment. Biol Cell 107:130–143
Wong GS et al (2018) Targeting wild-type KRAS-amplified gastroesophageal cancer through combined MEK and SHP2 inhibition. Nat Med 24:968–977
Xie J, Wang X, Proud CG (2016) mTOR inhibitors in cancer therapy. F1000Res 5:F1000
Xie C et al (2017) Identification of a New Potent Inhibitor Targeting KRAS in Non-small Cell Lung Cancer Cells. Front Pharmacol 8:823
Xiong Y et al (2017) Covalent guanosine mimetic inhibitors of G12C KRAS. ACS Med Chem Lett 8:61–66
Yamasaki K et al (1989) Conformation change of effector-region residues in antiparallel beta-sheet of human c-Ha-ras protein on GDP––GTP gamma S exchange: a two-dimensional NMR study. Biochem Biophys Res Commun 162:1054–1062
Yang MH et al (2012) Regulation of RAS oncogenicity by acetylation. Proc Natl Acad Sci USA 109:10843–10848
Yang JL et al (2016) A novel anti-p21Ras scFv antibody reacting specifically with human tumour cell lines and primary tumour tissues. BMC Cancer 16:131
Ye M et al (2005) Crystal structure of M-Ras reveals a GTP-bound "off" state conformation of Ras family small GTPases. J Biol Chem 280:31267–31275
Zhang Z, Shokat KM (2019) Bifunctional small-molecule ligands of K-Ras induce its association with immunophilin proteins. Angew Chem Int Ed Engl 58:16314–16319
Zhang J et al (2010) Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature 463:501–506
Zhang Y et al (2018) Design of small molecules that compete with nucleotide binding to an engineered oncogenic KRAS allele. Biochemistry 57:1380–1389
Zhou Y, Hancock JF (2015) Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta 1853:841–849
Zhou Y et al (2017) Lipid-sorting specificity encoded in K-Ras membrane anchor regulates signal output. Cell 168:239–251
Zimmermann G et al (2013) Small molecule inhibition of the KRAS-PDEdelta interaction impairs oncogenic KRAS signalling. Nature 497:638–642
Zimmermann G et al (2014) Structure guided design and kinetic analysis of highly potent benzimidazole inhibitors targeting the PDEdelta prenyl binding site. J Med Chem 57:5435–5448
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (410008598), the Canadian Cancer Society Research Institute (703209, 706696), Canada Foundation for Innovation (CFI), the Princess Margaret Cancer Foundation (PMCF) and Canada Research Chairs.
Author information
Authors and Affiliations
Contributions
All authors contributed to writing the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Marshall, C.B., KleinJan, F., Gebregiworgis, T. et al. NMR in integrated biophysical drug discovery for RAS: past, present, and future. J Biomol NMR 74, 531–554 (2020). https://doi.org/10.1007/s10858-020-00338-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-020-00338-6