Abstract
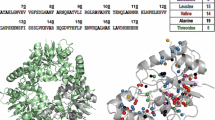
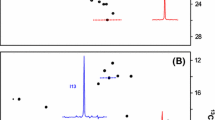
A new strategy for the NMR assignment of aliphatic side-chains in large perdeuterated proteins is proposed. It involves an alternative isotopic labeling protocol, the use of an out-and-back 13C–13C TOCSY experiment ((H)C-TOCSY-C-TOCSY-(C)H) and an optimized non-uniform sampling protocol. It has long been known that the non-linearity of an aliphatic spin-system (for example Ile, Val, or Leu) substantially compromises the efficiency of the TOCSY transfers. To permit the use of this efficient pulse scheme, a series of optimized precursors were designed to yield linear 13C perdeuterated side-chains with a single protonated CH3 group in these three residues. These precursors were added to the culture medium for incorporation into expressed proteins. For Val and Leu residues, the topologically different spin-systems introduced for the pro-R and pro-S methyl groups enable stereospecific assignment. All CH3 can be simultaneously assigned on a single sample using a TOCSY experiment. It only requires the tuning of a mixing delay and is thus more versatile than the relayed COSY experiment. Enhanced resolution and sensi-tivity can be achieved by non-uniform sampling combined with the removal of the large JCC coupling by deconvolution prior to the processing by iterative soft thresholding. This strategy has been used on malate synthase G where a large percentage of the CH3 groups could be correlated directly up to the backbone Ca. It is anticipated that this robust combined strategy can be routinely applied to large proteins.







Similar content being viewed by others
References
Ayala I, Hamelin O, Amero C et al (2012) An optimized isotopic labeling strategy of isoleucine-γ2 methyl groups for solution NMR studies of high molecular weight proteins. Chem Commun 48:1434. doi:10.1039/c1cc12932e
Bax A, Clore GM, Gronenborn AM (1990) 1H–1H correlation via isotropic mixing of 13C magnetization, a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J Magn Reson 88:425–431. doi:10.1016/0022-2364(90)90202-K
Braunschweiler L, Ernst RR (1983) Coherence transfer by isotropic mixing: application to proton correlation spectroscopy. J Magn Reson 53:521–528. doi:10.1016/0022-2364(83)90226-3
Chipman D, Barak Z, Schloss JV (1998) Biosynthesis of 2-aceto-2-hydroxy acids: acetolactate synthases and acetohydroxyacid synthases. Biochim Biophys Acta 1385:401–419. doi:10.1016/S0167-4838(98)00083-1
Davis DG, Bax A (1985) Assignment of complex proton NMR spectra via two-dimensional homonuclear Hartmann–Hahn spectroscopy. J Am Chem Soc 107:2820–2821. doi:10.1021/ja00295a052
Delaglio F, Grzesiek S, Vuister GW et al (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. doi:10.1007/BF00197809
Dumas R, Biou V, Halgand F et al (2001) Enzymology, structure, and dynamics of acetohydroxy acid isomeroreductase. Acc Chem Res 34:399–408. doi:10.1021/ar000082w
Eaton HL, Fesik SW, Glaser SJ, Drobny GP (1990) Time dependence of 13C–13C magnetization transfer in isotropic mixing experiments involving amino acid spin systems. J Magn Reson 90:452–463. doi:10.1016/0022-2364(90)90050-J
Eich G, Bodenhausen G, Ernst RR (1982) Exploring nuclear spin systems by relayed magnetization transfer. J Am Chem Soc 104:3731–3732. doi:10.1021/ja00377a036
Gans P, Hamelin O, Sounier R et al (2010) Stereospecific isotopic labeling of methyl groups for NMR spectroscopic studies of high-molecular-weight proteins. Angew Chem Int Ed 49:1958–1962. doi:10.1002/anie.200905660
Gardner KH, Kay LE (1997) Production and incorporation of 15N, 13C, 2H (1H-δ1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc 119:7599–7600. doi:10.1021/ja9706514
Godoy-Ruiz R, Guo C, Tugarinov V (2010) Alanine methyl groups as NMR probes of molecular structure and dynamics in high-molecular-weight proteins. J Am Chem Soc 132:18340–18350. doi:10.1021/ja1083656
Goto NK, Gardner KH, Mueller GA et al (1999) A robust and cost-effective method for the production of Val, Leu, Ile (δ1) methyl-protonated 15N-, 13C-, 2H-labeled proteins. J Biomol NMR 13:369–374. doi:10.1023/A:1008393201236
Güntert P, Braun W, Billeter M, Wüthrich K (2001) Automated stereospecific proton NMR assignments and their impact on the precision of protein structure determinations in solution. J Am Chem Soc 111:3997–4004. doi:10.1021/ja00193a036
Hill CM, Pang SS, Duggleby RG (1997) Purification of Escherichia coli acetohydroxyacid synthase isoenzyme II and reconstitution of active enzyme from its individual pure subunits. Biochem J 327:891–898. doi:10.1042/bj3270891
Hu W, Namanja AT, Wong S, Chen Y (2012) Selective editing of Val and Leu methyl groups in high molecular weight protein NMR. J Biomol NMR 53:113–124. doi:10.1007/s10858-012-9629-2
Hyberts SG, Märki W, Wagner G (1987) Stereospecific assignments of side-chain protons and characterization of torsion angles in Eglin c. Eur J Biochem 164:625–635. doi:10.1111/j.1432-1033.1987.tb11173.x
Kadkhodaie M, Rivas O, Tan M et al (1991) Broadband homonuclear cross polarization using flip-flop spectroscopy. J Magn Reson 91:437–443. doi:10.1016/0022-2364(91)90210-K
Kainosho M, Torizawa T, Iwashita Y et al (2006) Optimal isotope labeling for NMR protein structure determinations. Nature 440:52–57. doi:10.1038/nature04525
Kazimierczuk K, Orekhov VY (2011) Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed Engl 50:5556–5559. doi:10.1002/anie.201100370
Kerfah R, Plevin MJ, Pessey O et al (2014) Scrambling free combinatorial labeling of alanine-β, isoleucine-δ1, leucine-proS and valine-proS methyl groups for the detection of long range NOEs. J Biomol NMR 61:73–82. doi:10.1007/s10858-014-9887-2
Marion D (2010) Combining methods for speeding up multi-dimensional acquisition. Sparse sampling and fast pulsing methods for unfolded proteins. J Magn Reson 206:81–87. doi:10.1016/j.jmr.2010.06.007
Mas G, Crublet E, Hamelin O et al (2013) Specific labeling and assignment strategies of valine methyl groups for NMR studies of high molecular weight proteins. J Biomol NMR 57:251–262. doi:10.1007/s10858-013-9785-z
McCoy MA, Mueller L (1993) Selective decoupling. J Magn Reson Ser A 101:122–130. doi:10.1006/jmra.1993.1021
Neri D, Szyperski T, Otting G et al (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28:7510–7516. doi:10.1021/bi00445a003
Ollerenshaw JE, Tugarinov V, Skrynnikov NR, Kay LE (2005) Comparison of 13CH3, 13CH2D, and 13CHD2 methyl labeling strategies in proteins. J Biomol NMR 33:25–41. doi:10.1007/s10858-005-2614-2
Otten R, Chu B, Krewulak KD et al (2010) Comprehensive and cost-effective NMR spectroscopy of methyl groups in large proteins. J Am Chem Soc 132:2952–2960. doi:10.1021/ja907706a
Plevin MJ, Hamelin O, Boisbouvier J, Gans P (2011) A simple biosynthetic method for stereospecific resonance assignment of prochiral methyl groups in proteins. J Biomol NMR 49:61–67. doi:10.1007/s10858-010-9463-3
Pristovšek P, Franzoni L (2006) Stereospecific assignments of protein NMR resonances based on the tertiary structure and 2D/3D NOE data. J Comput Chem 27:791–797. doi:10.1002/jcc.20389
Rosen MK, Gardner KH, Willis RC et al (1996) Selective methyl group protonation of perdeuterated proteins. J Mol Biol 263:627–636. doi:10.1006/jmbi.1996.0603
Scholz I, Jehle S, Schmieder P et al (2007) J-deconvolution using maximum entropy reconstruction applied to 13C–13C solid-state cross-polarization magic-angle-spinning NMR of proteins. J Am Chem Soc 129:6682–6683. doi:10.1021/ja070849g
Senn H, Werner B, Messerle BA et al (1989) Stereospecific assignment of the methyl 1H NMR lines of valine and leucine in polypeptides by nonrandom 13C labeling. FEBS Lett 249:113–118. doi:10.1016/0014-5793(89)80027-4
Shaka AJ, Lee CJ, Pines A (1988) Iterative schemes for bilinear operators; application to spin decoupling. J Magn Reson 77:274–293. doi:10.1016/0022-2364(88)90178-3
Shimba N, Stern AS, Craik CS et al (2003) Elimination of 13Cα splitting in protein NMR spectra by deconvolution with maximum entropy reconstruction. J Am Chem Soc 125:2382–2383. doi:10.1021/ja027973e
Stern AS, Donoho DL, Hoch JC (2007) NMR data processing using iterative thresholding and minimum l(1)-norm reconstruction. J Magn Reson 188:295–300. doi:10.1016/j.jmr.2007.07.008
Tang C, Iwahara J, Clore GM (2005) Accurate determination of leucine and valine side-chain conformations using U-[15N/13C/2H]/[1H-(methine/methyl)-Leu/Val] isotope labeling, NOE pattern recognition, and methine Cγ–Hγ/Cβ–Hβ residual dipolar couplings: application to the 34-kDa Enzyme IIA Chitobiose. J Biomol NMR 33:105–121. doi:10.1007/s10858-005-1206-5
Tugarinov V, Kay LE (2003a) Ile, Leu, and Val methyl assignments of the 723-residue Malate Synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125:13868–13878. doi:10.1021/ja030345s
Tugarinov V, Kay LE (2003b) Side chain assignments of Ile-δ1 methyl groups in high molecular weight proteins: an application to a 46 ns tumbling molecule. J Am Chem Soc 125:5701–5706. doi:10.1021/ja021452
Tugarinov V, Kay LE (2004) Stereospecific NMR assignments of prochiral methyls, rotameric states and dynamics of valine residues in malate synthase G. J Am Chem Soc 126:9827–9836. doi:10.1021/ja048738u
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced 1H–13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428. doi:10.1021/ja030153x
Tugarinov V, Choy W-Y, Orekhov VY, Kay LE (2005) Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci USA 102:622–627. doi:10.1073/pnas.0407792102
Tugarinov V, Venditti V, Clore GM (2013) A NMR experiment for simultaneous correlations of valine and leucine/isoleucine methyls with carbonyl chemical shifts in proteins. J Biomol NMR 58:1–8. doi:10.1007/s10858-013-9803-1
Van Melckebeke H, Simorre J-P, Brutscher B (2004) Suppression of artifacts induced by homonuclear decoupling in amino-acid-type edited methyl 1H–13C correlation experiments. J Magn Reson 170:199–205. doi:10.1016/j.jmr.2004.06.016
Vuister GW, Yamazaki T, Torchia DA, Bax A (1993) Measurement of two- and three-bond 13C–1H J couplings to the C delta carbons of leucine residues in staphylococcal nuclease. J Biomol NMR 3:297–306. doi:10.1007/BF00212516
Wüthrich K, Wider G, Wagner G, Braun W (1982) Sequential resonance assignments as a basis for determination of spatial protein structures by high resolution proton nuclear magnetic resonance. J Mol Biol 155:311–319. doi:10.1016/0022-2836(82)90007-9
Acknowledgments
We would like to thank Dr. S.J. Remington for providing the MSG plasmid, Dr. D. Chipman for providing the ALSII plasmid, Pr. Vladislav Yu. Orekhov for providing MDDNMR software, Mrs I. Ayala, Drs. P. Gans and M. Plevin for stimulating discussions and Drs Laura Lerner and Michael Caffrey for manuscript editing. This work used the high-field NMR and the isotopic labeling facilities at the Grenoble Instruct Centre (ISBG; UMS 3518 CNRS-CEA-UJF-EMBL) with support from FRISBI (ANR-10-INSB-05-02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB). The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Program FP7/2007-2013 Grant Agreement No. 260887.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kerfah, R., Hamelin, O., Boisbouvier, J. et al. CH3-specific NMR assignment of alanine, isoleucine, leucine and valine methyl groups in high molecular weight proteins using a single sample. J Biomol NMR 63, 389–402 (2015). https://doi.org/10.1007/s10858-015-9998-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9998-4