Abstract
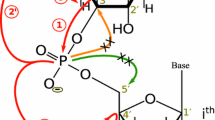
A fast, robust and reliable strategy for automated sequential resonance assignment for uniformly [13C, 15N]-labeled RNA via its phosphodiester backbone is presented. It is based on a series of high-dimensional through-bond APSY experiments: a 5D HCP-CCH COSY, a 4D H1′C1′CH TOCSY for ribose resonances, a 5D HCNCH for ribose-to-base connection, a 4D H6C6C5H5 TOCSY for pyrimidine resonances, and a 4D H8C8(C)C2H2 TOCSY for adenine resonances. The utilized pulse sequences are partially novel, and optimized to enable long evolution times in all dimensions. The highly precise APSY peak lists derived with these experiments could be used directly for reliable automated resonance assignment with the FLYA algorithm. This approach resulted in 98 % assignment completeness for all 13C–1H, 15N1/9 and 31P resonances of a stem-loop with 14 nucleotides.


Similar content being viewed by others
References
Aeschbacher T, Schubert M, Allain FH (2012) A procedure to validate and correct the 13C chemical shift calibration of RNA datasets. J Biomol NMR 52(2):179–190. doi:10.1007/s10858-011-9600-7
Aeschbacher T, Schmidt E, Blatter M, Maris C, Duss O, Allain FH, Güntert P, Schubert M (2013) Automated and assisted RNA resonance assignment using NMR chemical shift statistics. Nucleic Acids Res 41(18). doi:10.1093/nar/gkt665
Fürtig B, Richter C, Bermel W, Schwalbe H (2004) New NMR experiments for RNA nucleobase resonance assignment and chemical shift analysis of an RNA UUCG tetraloop. J Biomol NMR 28(1):69–79
Güntert P (2003) Automated NMR protein structure calculation. Prog Nucl Magn Reson Spectrosc 43(3–4):105–125. doi:10.1016/S0079-6565(03)00021-9
Hiller S, Fiorito F, Wüthrich K, Wider G (2005) Automated projection spectroscopy (APSY). Proc Natl Acad Sci USA 102(31):10876–10881. doi:10.1073/pnas.0504818102
IUPAC-IUB (1983) Abbreviations and symbols for the description of conformations of polynucleotide chains. Recommendations 1982. IUPAC-IUB Joint Commission on Biochemical Nomenclature (JCBN). Eur J Biochem FEBS 131(1):9–15
Kim S, Szyperski T (2003) GFT NMR, a new approach to rapidly obtain precise high-dimensional NMR spectral information. J Am Chem Soc 125(5):1385–1393. doi:10.1021/ja028197d
Krähenbühl B, Wider G (2012) Automated projection spectroscopy (APSY) for the assignment of NMR resonances in biological macromolecules. Chimia 66(10):767–771
Krähenbühl B, Hofmann D, Maris C, Wider G (2012) Sugar-to-base correlation in nucleic acids with a 5D APSY-HCNCH or two 3D APSY-HCN experiments. J Biomol NMR 52(2):141–150. doi:10.1007/s10858-011-9588-z
Kupce E, Freeman R (2003) Projection-reconstruction of three-dimensional NMR spectra. J Am Chem Soc 125(46):13958–13959. doi:10.1021/ja038297z
Lopez-Mendez B, Güntert P (2006) Automated protein structure determination from NMR spectra. J Am Chem Soc 128(40):13112–13122. doi:10.1021/ja061136l
Marino JP, Schwalbe H, Anklin C, Bermel W, Crothers DM, Griesinger C (1995) Sequential correlation of anomeric ribose protons and intervening phosphorus in RNA oligonucleotides by a 1H, 13C, 31P triple-resonance experiment—HCP-CCH-TOCSY. J Biomol NMR 5(1):87–92
Narayanan RL, Dürr UHN, Bibow S, Biernat J, Mandelkow E, Zweckstetter M (2010) Automatic assignment of the intrinsically disordered protein Tau with 441-residues. J Am Chem Soc 132(34):11906–11907. doi:10.1021/ja105657f
Nozinovic S, Fürtig B, Jonker HRA, Richter C, Schwalbe H (2010) High-resolution NMR structure of an RNA model system: the 14-mer cUUCGg tetraloop hairpin RNA. Nucleic Acids Res 38(2):683–694. doi:10.1093/Nar/Gkp956
Orekhov VY, Ibraghimov I, Billeter M (2003) Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR 27(2):165–173. doi:10.1023/A:1024944720653
Pardi A, Douglas JT, Latham MP, Armstrong GS, Bendiak B (2008) High-resolution pyrimidine- and ribose-specific 4D HCCH-COSY spectra of RNA using the filter diagonalization method. J Biomol NMR 41(4):209–219. doi:10.1007/s10858-008-9253-3
Ramachandran R, Sich C, Grüne M, Soskic V, Brown LR (1996) Sequential assignments in uniformly 13C- and 15N-labelled RNAs: The HC(N, P) and HC(N, P)-CCH-TOCSY experiments. J Biomol NMR 7(3):251–255
Schmidt E, Güntert P (2012) A new algorithm for reliable and general NMR resonance assignment. J Am Chem Soc 134(30):12817–12829. doi:10.1021/Ja305091n
Tate S, Ono A, Kainosho M (1995) Sequential backbone assignment in 13C-labeled DNA by the 1H, 13C, 31P triple-resonance experiment, HCP-CCH-COSY. J Magn Reson Ser B 106(1):89–91. doi:10.1006/jmrb.1995.1016
Acknowledgments
The Swiss National Science Foundation (SNSF project 200021_120048) and the Institute for Molecular Biology and Biophysics (ETH Zurich) are gratefully acknowledged for financial support. P.G. gratefully acknowledges financial support by the Lichtenberg program of the Volkswagen Foundation and the Japan Society for the Promotion of Science (JSPS).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Krähenbühl, B., El Bakkali, I., Schmidt, E. et al. Automated NMR resonance assignment strategy for RNA via the phosphodiester backbone based on high-dimensional through-bond APSY experiments. J Biomol NMR 59, 87–93 (2014). https://doi.org/10.1007/s10858-014-9829-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-014-9829-z