Abstract
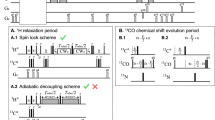
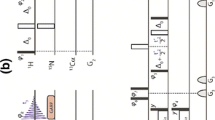
Unprotected amide protons can undergo fast hydrogen exchange (HX) with protons from the solvent. Generally, NMR experiments using the out-and-back coherence transfer with amide proton detection are affected by fast HX and result in reduced signal intensity. When one of these experiments, 1H–15N HSQC, is used to measure the 15N transverse relaxation rate (R2), the measured R2 rate is convoluted with the HX rate (kHX) and has higher apparent R2 values. Since the 15N R2 measurement is important for analyzing protein backbone dynamics, the HX effect on the R2 measurement is investigated and described here by multi-exponential signal decay. We demonstrate these effects by performing 15N R CPMG2 experiments on α-synuclein, an intrinsically disordered protein, in which the amide protons are exposed to solvent. We show that the HX effect on R CPMG2 can be extracted by the derived equation. In conclusion, the HX effect may be pulse sequence specific and results from various sources including the J coupling evolution, the change of steady state water proton magnetization, and the D2O content in the sample. To avoid the HX effect on the analysis of relaxation data of unprotected amides, it is suggested that NMR experimental conditions insensitive to the HX should be considered or that intrinsic R CPMG2 values be obtained by methods described herein.






Similar content being viewed by others
References
Bertini I, Carrano CJ, Luchinat C, Piccioli M, Poggi L (2002) A 15N NMR mobility study on the dicalcium P43 M calbindin D9 k and its mono-La3 + -substituted form. Biochemistry 41:5104–5111
Bertoncini CW, Rasia RM, Lamberto GR, Binolfi A, Zweckstetter M, Griesinger C, Fernandez CO (2007) Structural characterization of the intrinsically unfolded protein beta-synuclein, a natural negative regulator of alpha-synuclein aggregation. J Mol Biol 372:708–722
Buevich AV, Baum J (1999) Dynamics of unfolded proteins: incorporation of distributions of correlation times in the model free analysis of NMR relaxation data. J Am Chem Soc 121:8671–8672
Buevich AV, Shinde UP, Inouye M, Baum J (2001) Backbone dynamics of the natively unfolded pro-peptide of subtilisin by heteronuclear NMR relaxation studies. J Biomol NMR 20:233–249
Bussell R Jr, Eliezer D (2001) Residual structure and dynamics in Parkinson’s disease-associated mutants of alpha-synuclein. J Biol Chem 276:45996–46003
Chen J, Tjandra N (2011) Water proton spin saturation affects measured protein backbone 15N spin relaxation rates. J Magn Reson 213:151–157
Chevelkov V, Xue Y, Rao DK, Forman-Kay JD, Skrynnikov NR (2010) 15NH/D-SOLEXSY experiment for accurate measurement of amide solvent exchange rates: application to denatured drkN SH3. J Biomol NMR 46:227–244
Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366
Croke RL, Sallum CO, Watson E, Watt ED, Alexandrescu AT (2008) Hydrogen exchange of monomeric alpha-synuclein shows unfolded structure persists at physiological temperature and is independent of molecular crowding in Escherichia coli. Protein Sci 17:1434–1445
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dempsey CE (2001) Hydrogen exchange in peptides and proteins using NMR spectroscopy. Prog Nucl Magn Reson Spectrosc 39:135–170
Eliezer D (2009) Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol 19:23–30
Eliezer D, Kutluay E, Bussell R Jr, Browne G (2001) Conformational properties of alpha-synuclein in its free and lipid-associated states. J Mol Biol 307:1061–1073
Farrow NA, Muhandham R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE (1994) Backbone dynamics of a free and a phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33:5984–6003
Gemmecker G, Jahnke W, Kessler H (1993) Measurement of fast proton exchange rates in isotopically labeled compounds. J Am Chem Soc 115:11620–11621
Goedert M (2001) Alpha-synuclein and neurodegenerative diseases. Nat Rev Neurosci 2:492–501
Griesinger C, Ernst RR (1988) Cross relaxation in time-dependent nuclear spin systems: invariant trajectory approach. Chem Phys Lett 152:239–247
Hansen DF, Led JJ (2003) Implications of using approximate Bloch–McConnell equations in NMR analyses of chemically exchanging systems: application to the electron self-exchange of plastocyanin. J Magn Reson 163:215–227
Hwang TL, van Zijl PC, Mori S (1998) Accurate quantitation of water-amide proton exchange rates using the phase-modulated CLEAN chemical EXchange (CLEANEX-PM) approach with a Fast-HSQC (FHSQC) detection scheme. J Biomol NMR 11:221–226
Iwahara J, Jung YS, Clore GM (2007) Heteronuclear NMR spectroscopy for lysine NH3 groups in proteins: unique effect of water exchange on 15N transverse relaxation. J Am Chem Soc 129:2971–2980
Kateb F, Pelupessy P, Bodenhausen G (2007) Measuring fast hydrogen exchange rates by NMR spectroscopy. J Magn Reson 184:108–113
Kay LE, Torchia DA, Bax A (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972–8979
Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE (2002) An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. J Am Chem Soc 124:10743–10753
Mandel AM, Akke M, Palmer AG 3rd (1995) Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol 246:144–163
McConnell HM (1958) Reaction rates by nuclear magnetic resonance. J Chem Phys 28:430–431
McNulty BC, Tripathy A, Young GB, Charlton LM, Orans J, Pielak GJ (2006) Temperature-induced reversible conformational change in the first 100 residues of alpha-synuclein. Protein Sci 15:602–608
Mittag T, Forman-Kay JD (2007) Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol 17:3–14
Mittermaier AK, Kay LK (2009) Observing biological dynamics at atomic resolution using NMR. Trends Biochem Sci 34:601–611
Palmer AG III (2004) NMR characterization of the dynamics of biomacromolecules. Chem Rev 104:3623–3640
Palmer AG III, Skelton NJ, Chazin WJ, Wright PE, Rance M (1992) Suppression of the effects of cross-correlation between dipolar and anisotropic chemical shift relaxation mechanisms in the measurement of spin–spin relaxation rates. Mol Phys 75:699–711
Palmer AG III, Kroenke CD, Loria JP (2001) Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Method Enzymol 339:204–238
Skrynnikov NR, Ernst RR (1999) Detection of intermolecular chemical exchange through decorrelation of two-spin order. J Magn Reson 137:276–280
Tugarinov V, Kay LE (2003) Quantitative NMR studies of high molecular weight proteins: application to domain orientation and ligand binding in the 723 residue enzyme malate synthase G. J Mol Biol 327:1121–1133
Uversky VN, Gillespie JR, Fink AL (2000) Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 41:415–427
Wang C, Palmer AG 3rd (2003) Solution NMR methods for quantitative identification of chemical exchange in 15N-labeled proteins. Magn Reson Chem 41:866–876
Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT Jr (1996) NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry 35:13709–13715
Wright PE, Dyson HJ (2009) Linking folding and binding. Curr Opin Struct Biol 19:31–38
Wu K-P, Kim S, Fela DA, Baum J (2008) Characterization of conformational and dynamic properties of natively unfolded human and mouse alpha-synuclein ensembles by NMR: implication for aggregation. J Mol Biol 378:1104–1115
Wu K-P, Weinstock DS, Narayanan C, Levy RM, Baum J (2009) Structural reorganization of alpha-synuclein at low pH observed by NMR and REMD simulations. J Mol Biol 391:784–796
Zhang YZ, Paterson Y, Roder H (1995) Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein Sci 4:804–814
Acknowledgments
K-PW was supported by an NIH Interdisciplinary Research Workforce Fellowship (5R90DK071502). This work has been supported by NIH grants (GM087012 and GM45302) and NSF grants (DBI-0403062 and DBI-0320746) to JB.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, S., Wu, KP. & Baum, J. Fast hydrogen exchange affects 15N relaxation measurements in intrinsically disordered proteins. J Biomol NMR 55, 249–256 (2013). https://doi.org/10.1007/s10858-013-9706-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-013-9706-1