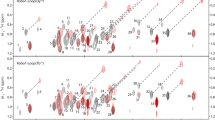
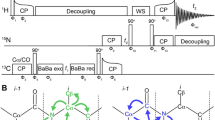
Abstract
New 13C-detected NMR experiments have been devised for molecules in solution and solid state, which provide chemical shift correlations of methyl groups with high resolution, selectivity and sensitivity. The experiments achieve selective methyl detection by exploiting the one bond J-coupling between the 13C-methyl nucleus and its directly attached 13C spin in a molecule. In proteins such correlations edit the 13C-resonances of different methyl containing residues into distinct spectral regions yielding a high resolution spectrum. This has a range of applications as exemplified for different systems such as large proteins, intrinsically disordered polypeptides and proteins with a paramagnetic centre.







Similar content being viewed by others
References
Amero C, Schanda P, Durá MA, Ayala I, Marion D, Franzetti B, Brutscher B, J Boisbouvier (2009) Fast two-dimensional NMR spectroscopy of high molecular weight protein assemblies. J Am Chem Soc 131:3448–3449
Andronesi OC, Becker S, Seidel K, Heise H, Young HS, Baldus M (2005) Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J Am Chem Soc 127:12965–12974
Arnesano F (2010) The role of copper ion and the ubiquitin system in neurodegenerative disorders. Ideas in chemistry and molecular sciences. Wiley, New York, pp 1–30
Arnesano F, Scintilla S, Calò V, Bonfrate E, Ingrosso C, Losacco M, Pellegrino T, Rizzarelli E, Natile G (2009) Copper-triggered aggregation of Ubiquitin. PLoS One 4:e7052
Atreya HS, Chary KVR (2001) Selective ‘unlabeling’ of amino acids in fractionally C-13 labeled proteins: an approach for stereospecific NMR assignments of CH3 groups in Val and Leu residues. J Biomol NMR 19:267–272
Barnwal RP, Atreya HS, Chary KVR (2008) Chemical shift based editing of CH3 groups in fractionally C-13-labelled proteins using GFT (3,2)D CT-HCCH-COSY: stereospecific assignments of CH3 groups of Val and Leu residues. J Biomol NMR 42:149–154
Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K (1995) The program xeasy for computer-supported NMR spectral-analysis of biological macromolecules. J Biomol NMR 6:1–10
Bennett AE, Rienstra CM, Auger M, Lakshmi KV, Griffin RG (1995) Heteronuclear decoupling in rotating solids. J Chem Phys 103:6951–6958
Bermel W, Bertini I, Felli IC, Piccioli M, Pierattelli R (2006) C-13-detected protonless NMR spectroscopy of proteins in solution. Prog Nucl Magn Reson Spectrosc 48:25–45
Bermel W, Bertini I, Felli IC, Matzapetakis M, Pierattelli R, Theil EC, Turano P (2007) A method for Cα direct-detection in protonless NMR. J Magn Reson 188:301–310
Bermel W, Felli IC, Kummerle R, Pierattelli R (2008) C-13 direct-detection biomolecular NMR. Concepts Magn Reson 32A:183–200
Bertini I, Luchinat C, Parigi G (2002) Paramagnetic constraints: an aid for quick solution structure determination of paramagnetic metalloproteins. Concepts Magn Reson 14:259–286
Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ (2007) Protein NMR spectroscopy. Academic Press, San Diego
Chandra K, Jaipuria G, Shet D, Atreya HS (2012) Efficient sequential assignments in proteins with reduced dimensionality 3D HN(CA)NH. J Biomol NMR 52:115–126
Chen L, Kaiser JM, Polenova T, Yang J, Rienstra CM, Müller LJ (2007a) Backbone assignments in solid-state proteins using J-based 3D Heteronuclear correlation spectroscopy. J Am Chem Soc 129:10650–10651
Chen L, Kaiser JM, Lai JF, Polenova T, Yang J, Rienstra CM, Müller LJ (2007b) J-based 2D homonuclear and heteronuclear correlation in solid-state proteins. Magn Reson Chem 45:S84–S92
Choy WY, Kay LE (2003) Model selection for the interpretation of protein side chain methyl dynamics. J Biomol NMR 25:325–333
Clore GM, Iwahara J (2009) Theory, practice, and applications of paramagnetic relaxation enhancement for the characterization of transient low-population states of biological macromolecules and their complexes. Chem Rev 109:4108–4139
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPIPE—a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293
Elena B, Lesage A, Steuernagel S, Bockmann A, Emsley L (2005) Proton to carbon-13 INEPT in solid-state NMR spectroscopy. J Am Chem Soc 127:17296–17302
Gagne SM, Tsuda S, Spyracopoulos L, Kay LE, Sykes BD (1998) Backbone and methyl dynamics of the regulatory domain of troponin C: anisotropic rotational diffusion and contribution of conformational entropy to calcium affinity. J Mol Biol 278:667–686
Gardner KH, Konrat R, Rosen MK, Kay LE (1996) An (H)C(CO)NH-TOCSY pulse scheme for sequential assignment of protonated methyl groups in otherwise deuterated N-15, C-13-labeled proteins. J Biomol NMR 8:351–356
Gardner KH, Rosen MK, Kay LE (1997) Global folds of highly deuterated, methyl-protonated proteins by multidimensional NMR. Biochemistry 36:1389–1401
Gross JD, Gelev VM, Wagner G (2003) A sensitive and robust method for obtaining intermolecular NOEs between side chains in large protein complexes. J Biomol NMR 25:235–242
Iwahara J, Tang C, Clore GM (2007) Practical aspects of (1)H transverse paramagnetic relaxation enhancement measurements on macromolecules. J Magn Reson 184:185–195
Jaipuria G, Thakur A, D’Silva P, Atreya HS (2010) High-resolution methyl edited GFT NMR experiments for protein resonance assignments and structure determination. J Biomol NMR 48:137–145
Jordan JB, Kovacs H, Wang Y, Mobli M, Luo R, Anklin C, Hoch JC, Kriwacki RW (2006) Three-dimensional 13C-detected CH3-TOCSY using selectively protonated proteins: facile methyl resonance assignment and protein structure determination. J Am Chem Soc 128:9119–9128
Kovacs H, Moskau D, Spraul M (2005) Cryogenically cooled probes—a leap in NMR technology. Prog Nucl Magn Reson Spectrosc 46:131–155
Metz G, Wu XL, Smith SO (1994) Ramped-amplitude cross polarization in magic-angle-spinning NMR. J Magn Reson, Series A 110:219–227
Metzler WJ, Wittekind M, Goldfarb V, Mueller L, Farmer BT (1996) Incorporation of H-1/C-13/N-15-{Ile, Leu, Val} into a perdeuterated, N-15-labeled protein: potential in structure determination of large proteins by NMR. J Am Chem Soc 118:6800–6801
Milardi D, Arnesano F, Grasso G, Magrì A, Tabbì G, Scintilla S, Natile G, Rizzarelli E (2007) Ubiquitin stability and the Lys 63-linked polyubiquitination site are compromised on copper binding. Angew Chem Int Ed 46:7993–7995
Nicholson LK, Kay LE, Baldisseri DM, Arango J, Young PE, Bax A, Torchia DA (1992) Dynamics of methyl-groups in proteins as studied by proton-detected C-13 NMR-spectroscopy—application to the leucine residues of staphylococcal nuclease. Biochemistry 31:5253–5263
Ottiger M, Delaglio F, Bax A (1998) Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra. J Magn Reson 131:373–378
Rosen MK, Gardner KH, Willis RC, Parris WE, Pawson T, Kay LE (1996) Selective methyl group protonation of perdeuterated proteins. J Mol Biol 263:627–636
Swain M, Slomiany MG, Rosenzweig SA, Atreya HS (2010) High-yield bacterial expression and structural characterization of recombinant human insulin-like growth factor binding protein-2. Arch Biochem Biophys 501:195–200
Takegoshi K, Nakamura S, Terao T (2001) C-13-H-1 dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem Phys Lett 344:631–637
Takegoshi K, Nakamura S, Terao T (2003) C-13-H-1 dipolar-driven C-13-C-13 recoupling without C-13 rf irradiation in nuclear magnetic resonance of rotating solids. J Chem Phys 118:2325–2341
Thruber KR, Tycko R (2009) Measurement of sample temperature under magic angle spinning from the chemical shift and spin lattice relaxation rate of 79Br in KBr powder. J Magn Reson 196:84–87
Tian Y, Chen LL, Niks D, Kaiser JM, Lai JF, Rienstra CM, Dunn MF, Mueller LJ (2009) J-Based 3D sidechain correlation in solid-state proteins. Phys Chem Chem Phys 11:7078–7086
Tugarinov V, Kay LE (2004) An isotope labeling strategy for methyl TROSY spectroscopy. J Biomol NMR 28:165–172
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced H-1-C-13 NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428
Tugarinov V, Hwang PM, Kay LE (2004) Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Ann Rev Biochem 73:107–146
Tugarinov V, Choy WY, Orekhov VY, Kay LE (2005) Solution NMR-derived global fold of a monomeric 82-kDa enzyme. Proc Natl Acad Sci USA 102:622–627
Werbelow LG, Marshall AG (1973) Internal-rotation and nonexponential methyl nuclear relaxation for macromolecules. J Magn Reson 11:299–313
Zech SG, Olejniczak E, Hajduk P, Mack J, McDermott AE (2004) Characterization of protein—ligand interactions by high-resolution solid-state NMR spectroscopy. J Am Chem Soc 126:13948–13953
Zheng DY, Huang YPJ, Moseley HNB, Xiao R, Aramini J, Swapna GVT, Montelione GT (2003) Automated protein fold determination using a minimal NMR constraint strategy. Protein Sci 12:1232–1246
Zhong L, Bamm VV, Ahmed MAM, Harauz G, Ladizhansky V (2007) Solid-state NMR spectroscopy of 18.5 kDa myelin basic protein reconstituted with lipid vesicles: spectroscopic characterisation and spectral assignments of solvent-exposed protein fragments. Biochim Biophys Acta Biomembranes 1768:3193–3205
Zwahlen C, Gardner KH, Sarma SP, Horita DA, Byrd RA, Kay LE (1998) An NMR experiment for measuring methyl–methyl NOEs in C-13-labeled proteins with high resolution. J Am Chem Soc 120:7617–7625
Acknowledgments
The facilities provided by NMR Research Centre at IISc supported by Department of Science and Technology (DST), India is gratefully acknowledged. HSA acknowledges support from Indo-Australia biotechnology fund awarded by the Department of Biotechnology (DBT). GJ acknowledges fellowship from Council of Scientific and Industrial Research (CSIR). The authors are sincerely thankful to Dr. Peter Güntert for useful discussions on structure calculation of Cu(II)-ubiquitin using CYANA. We thank Dr. John Cort, Pacific Northwest National Laboratory, for providing the Ubiquitin plasmid.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jaipuria, G., Lobo, N.P., Shet, D. et al. High resolution methyl selective 13C-NMR of proteins in solution and solid state. J Biomol NMR 54, 33–42 (2012). https://doi.org/10.1007/s10858-012-9647-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-012-9647-0