Abstract
The known biological functions of RNA have expanded in recent years and now include gene regulation, maintenance of sub-cellular structure, and catalysis, in addition to propagation of genetic information. As for proteins, RNA function is tightly correlated with structure. Unlike proteins, structural information for larger, biologically functional RNAs is relatively limited. NMR signal degeneracy, relaxation problems, and a paucity of long-range 1H–1H dipolar contacts have limited the utility of traditional NMR approaches. Selective isotope labeling, including nucleotide-specific and segmental labeling strategies, may provide the best opportunities for obtaining structural information by NMR. Here we review methods that have been developed for preparing and purifying isotopically labeled RNAs, as well as NMR strategies that have been employed for signal assignment and structure determination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
RNA molecules play pivotal roles in a large and growing number of known biological functions. Initially relegated to roles in genetic information transfer, RNA species comprising as few as ~19 nucleotides (nt), or as many as ~4,700 nt or more, are now known to play roles in transcriptional regulation, metabolite and protein recognition, catalysis, maintenance of sub-cellular and viral structure, and other essential functions (Hassouna et al. 1984; Doudna and Rath 2002; Bartel 2004; Kim 2005; Brodersen and Voinnet 2006; Boisvert et al. 2007; Edwards et al. 2007; Korostelev and Noller 2007; Wakeman et al. 2007; Bessonov et al. 2008; Steitz 2008). In addition, a significant portion of the transcribed Eukaryotic genome produces non-protein-coding RNAs, of which only a few have known biological roles (Ponting et al. 2009). Current understanding at the atomic level of how RNAs function is generally limited, a problem that is compounded by the relative paucity of high-resolution structural information. To date, only ~1,300 RNA structures have been deposited in Nucleic Acid Database (NDB), whereas, more than 55,000 protein structures have been deposited in the Protein Databank (PDB). Conformational heterogeneity and a relatively uniform, negative surface charge are two significant impediments to structural studies by X-ray crystallography.
NMR is a potentially powerful tool for studying the structure and dynamics of RNA (Wüthrich 1986). Unfortunately, because most RNAs contain only four types of residues, chemical shift dispersion of 1H, 13C, 15N and 31P nuclei is relatively low. Overall proton density is also much lower than that of proteins, and non-exchangeable proton density is concentrated mainly within the major grooves of A-helices. In addition, interproton distances between different secondary structure elements are typically greater than 5 Å, which limits the utility of NOE experiments for establishing overall RNA folds (Allain and Varani 1997; Lukavsky and Puglisi 2005). Finally, NMR assignments are critically dependent on analysis of signals for aromatic C–H groups, and 1H–13C dipolar coupling can severely limit the sensitivity and resolution of 1H–13C correlation NMR spectra obtained for these groups in larger RNAs with longer rotational correlation times. For these reasons, high-resolution NMR-based structural studies have been applied mainly to relatively small RNAs: Of the 298 RNA NMR structures that have been deposited to date in the Nucleic Acid Database, only three are comprised of 50 or more nucleotides.
The study of RNA structure has typically relied on the “divide-and-conquer” strategy, in which functional RNAs are dissected into smaller sub-domains (for examples, see: (Aboul-ela et al. 1996; Amarasinghe et al. 2000; Spriggs et al. 2008). Unfortunately, the isolated sub-domains typically lack function, and it is not always clear that their folding matches the folding that occurs in the context of the intact RNA. In fact, examples have emerged in which the isolated and intact structures differ significantly (Abramovitz and Pyle 1997). Structures of small RNAs can also be susceptible to crystal packing effects (Shah and Brunger 1999).
Nucleotide specific 13C, 15N, and 2H isotope labeling was introduced in the early 90 s as a means of overcoming signal degeneracy problems (Batey et al. 1992, 1995; Nikonowicz and Pardi 1992; Michnicka et al. 1993; Kim et al. 1995). More recently, segmental labeling approaches have been developed for studying sub-domain structure in the context of intact, functional RNAs (Xu and Crothers 1996; Kim et al. 2002; Lukavsky et al. 2003). This strategy should work well for RNAs with subdomains that have relatively short rotational correlation times, and could be applicable to RNAs with longer rotational correlation times using appropriate TROSY-based NMR experiments (Pervushin et al. 1997; Riek et al. 1999; Fiala et al. 2000). This review outlines the methods that have been developed for the preparation and purification of milligram quantities of isotopically-labeled RNAs, as well as the NMR assignment strategies that could eventually allow high resolution structure determination of RNA subdomains within very large (>700 nt) RNAs.
RNA synthesis
Chemical synthesis
Chemical synthesis of RNA became commercially feasible in the late 80’s with the development of efficient ribose 2′-hydroxyl protecting groups (Usman et al. 1987; Scaringe et al. 1998). Although modified NTPs can readily be incorporated into specific positions, low coupling efficiencies limit this approach to relatively small RNAs. At present, chemical synthesis is practical for RNAs comprising fewer than ~55 nt. Site-specific labeling of even these relatively small RNAs could be extremely useful for incorporation into larger RNAs by enzymatic ligation (see below). Unfortunately, isotopically-labeled phosphoramidites for RNA synthesis are not commercially available.
Enzymatic synthesis using T7 RNA polymerase
RNAs for NMR-based structure determination are now commonly prepared by in vitro transcription using the T7 RNA polymerase (RNAP) enzyme. Milligram quantities of RNAs ranging in length from 10 to 30,000 nt can be prepared, and it is straightforward to incorporate commercially available 13C-, 15N-, and 2H-isotopically labeled nucleotides (Milligan and Uhlenbeck 1989; Pokrovskaya and Gurevich 1994). T7 RNAP is highly specific for its own promoters and exhibits no affinity for the closely related T3 promoters. The error frequency in transcripts for wild type T7 RNA polymerase is as low as 6 × 10−5.
Although T7 RNAP-dependent in vitro transcription is currently the most popular and widely used method for preparing RNAs, the approach does have some drawbacks. First, to achieve the best yields, the most commonly used T7 class III promoter requires guanosine at the first and second positions of the sequence. As such, non-native 5′-guanosines sometimes must be added to the RNA sequence of interest. Similarly, the T7 Φ2.5 class II promoter requires a 5′-AG sequence for efficient transcription. These 5′-sequence requirements can be overcome by engineering the appropriate 5′-GG (or 5′-AG) start sequence into a hammerhead (HH) ribozyme sequence, which itself is appended to the 5′ end of the fully native target RNA sequence (discussed further below).
A second drawback is that T7 RNAP often adds one or more non-templated nucleotides (typically Cs) to the 3′-terminus of the product RNA (Milligan et al. 1987; Milligan and Uhlenbeck 1989; Pokrovskaya and Gurevich 1994). Although 3′-heterogeneity is not a significant problem for most structural studies, it can be a major problem if the RNA is to be ligated to the 5′-end of a second RNA molecule. 5′-end heterogeneity can also be a problem for transcripts that have more than 3 consecutive 5’-guanosines (Pleiss et al. 1998), or for constructs with other 5′-terminal sequences such as 5′-CACUGU, 5′-CAGAGA or 5′-GAAAAA (Helm et al. 1999). 5′-end heterogeneity does not appear to be a problem when using the T7 Φ2.5 class II promoter.
Control of 5′-end homogeneity
The efficient way to produce RNA transcripts with uniform 5′-ends is to construct a DNA template with a self-cleaving hammerhead sequence immediately upstream of the RNA sequence of interest. The HH ribozyme is a relatively small structural element ( ~50 nt) that catalyzes phosphodiester bond hydrolysis specifically at its 3′-GUC sequence. The HH ribozyme folds into the catalytically active structure immediately upon transcription and self-cleaves in the presence of Mg2+. The desired RNA product contains a 5′ hydroxyl group which, as discussed below, can be beneficial for subsequent ligation (Fig. 1a) (Birikh et al. 1997). Another advantage of using a 5′ hammerhead cassette is that the optimized transcription start sequence (e.g., 5′-GG for the T7 class III promoter) can be incorporated into the ribozyme sequence, rather than the RNA of interest.
Method used to produce RNA with homogeneous 5′ and 3′ ends. a Hammerhead (HH) cleavage produces homogeneous 5′-end; b HDV/HH ribozyme, RNase P cleavage or 2′-O-Me of the last two nucleotides on the DNA template can produce 3′-homogeneous ends. c RNase H cleavage of the transcribed RNA can produce both homogeneous 5′ and 3′ ends at the cleaving point. For details see Hartmann et al. (2005)
Control of 3′ end homogeneity
Several methods are presently available for controlling 3′ end homogeneity. For smaller RNAs (<60 nt) that can be generated from synthetic templates, non-templated 3′ nucleotide addition can be inhibited by incorporating two 2′-O-methoxyribose-modified nucleotides (commercially available) at the 5′-end of the template strand (Kao et al. 1999) (Fig. 1b). For larger RNAs, the DNA template can be modified such that it encodes for a HH ribozyme at the 3′-end of the RNA of interest. However, HH ribozymes preferentially cleave at sites that contain a 3′-GUC sequence, and this would restrict sequence options for the 3′-end of the desired RNA.
The Hepatitis delta virus (HDV) ribozyme has no such sequence requirements and can be used to control 3′ end homogeneity of any RNA sequence of interest. The HDV ribozyme is active during transcription in the presence of Mg2+. The product RNA contains a 2′,3′-cyclic phosphate at its 3′ end (Fig. 1b), which can be advantageous for controlling subsequent ligation reactions (see below). A frequently observed problem is that the upstream RNA sequence can interfere with the proper folding of the HDV ribozyme, leading to poor cleavage efficiency. This problem can sometimes be overcome by cycling the temperature to refold the RNA, which allows the ribozyme domain to adopt the active structure (at least transiently) for self-cleavage. HDV cleavage efficiency can also be improved by addition of Ca2+ (rather than Mg2+) as a cofactor (Cerrone-Szakal et al. 2009). An additional approach we have used is to redesign the HDV ribozyme so that it does not form secondary structures with the appended RNA of interest, based on MFOLD predictions (Zuker 2003).
The RNase P ribozyme can also be employed to control 3′-end homogeneity. In this approach, a tRNA sequence is appended to the 3′-end of the desired RNA sequence. RNase P recognizes and cleaves the 5′-end of the tRNA, affording the product RNA of interest. Like HDV, RNase P does not have specific sequence requirements for residues upstream of the cleavage site. The major advantage of this method over HDV-dependent cleavage is that there is generally no folding inference, due to the fact that tRNAs adopt highly stable, autonomous structures. The product RNAs contain 3′-hydroxyl termini (Fig. 1b), in contrast to the 2′, 3′-cyclic phosphates generated by HDV and HH cleavage, which is suitable for many downstream applications.
Control of both 5′ and 3′ end homogeneity
RNase H can recognize and bind to DNA/RNA hybrid duplexes and cleave the backbone of the RNA, leaving a 5′-phosphate and 3′-hydroxyl group (Berkower and Leis 1973) (Fig. 1c). This enzyme can therefore be used to control homogeneity at both the 5′ and 3′ ends of the target RNA. When the target RNA and an oligonucleotide containing a short stretch of DNA flanked by 2′-O-Me RNA sequences form a hybrid duplex (Fig. 1c), the site of cleavage is specific but apparently dependent on the commercial source of the RNase H (Inoue et al. 1987; Hartmann et al. 2005). RNase H can be used to remove non-native 5′-elements that contain optimized T7 transcription start sites (Lapham and Crothers 1996).
RNA purification methods
Purification of large quantities of RNAs is typically achieved by denaturing polyacrylamide gel electrophoresis (PAGE; 13 by 16.5′′ gel box size) (Wyatt et al. 1991). This is a robust method that works well for a wide range of RNA sizes. Nucleotide-level resolution can typically be achieved for RNAs of up to ~30 residues. This method can be laborious, and often requires the use of several gels per NMR sample. The standard procedure includes an initial ethanol precipitation step, which in our hands often leads to the formation of insoluble RNA aggregates. We typically skip this step, and directly add the transcription reaction mixture to the gel. For RNAs that contain self-complementary dimerization elements, best results have been obtained by running the gels in a warm environment (e.g., at ~50°C in a small enclosed area with a space heater). This is necessary because RNA fragments resulting from aborted synthesis can associate with the full-length construct of interest if both contain the self-complementary dimerization sequence. Ion-pair reversed-phase HPLC and anion-exchange HPLC have also been employed to purify RNA under denaturing conditions (Anderson et al. 1996; Shields et al. 1999; Azarani and Hecker 2001). These approaches are much less laborious and time-consuming, but the best results have been achieved when applied to relatively small RNAs. Of course, purification under denaturing conditions requires desalting and refolding of the RNA prior to structural studies.
Several additional chromatographic methods have been developed for purifying RNA under non-denaturing conditions, including size exclusion (Lukavsky and Puglisi 2004; Kim et al. 2007; McKenna et al. 2007), DNA-affinity (Cheong et al. 2004), and protein-affinity (Crowe et al. 1994; Batey and Kieft 2007) column chromatography (for a recent review see (Dayie 2008)). An affinity-tag based purification procedure was also developed recently, in which protein-RNA interactions immobilize RNAs carrying both a modified HDV sequence and a 3′-affinity tag (Batey and Kieft 2007). After the immobilization, the target RNA is cleaved by the addition of imidazole, which activates the modified ribozyme. This purification approach also affords product RNAs with homogeneous 3′-ends.
Isotope labeling strategies
Residue-specific isotopic labeling has greatly facilitated structural studies of large proteins and protein assemblies by reducing NMR spectral complexity and unfavorable relaxation behavior (Fiaux et al. 2004; Sprangers and Kay 2007). Proteins enriched with 2H, 15N and/or 13C can be directly prepared in E. Coli by commercially available materials (Sprangers and Kay 2007), and site-specifically labeled proteins can be synthesized by cell-free methods (Kainosho et al. 2006). A variety of site-specifically deuterated and isotopically labeled amino acids are currently available for amino acid-specific labeling. In comparison, relatively few reagents are commercially available for the preparation of isotopically labeled RNAs. Until very recently, it has only been possible to obtain uniformly enriched rNTPs (13C, 15N, 13C/15N, or 2H) from commercial sources (and at considerable expense).
Ribose labeling
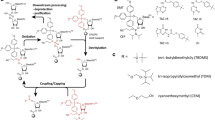
Ribonucleotide triphosphates that contain 2H and/or 2H, 13C specifically labeled ribose moieties can be prepared enzymatically, but the process is laborious and requires more than a dozen enzymatic reactions (Fig. 2) (Scott et al. 2000). Procedures have been developed by Williamson and co-workers to prepare variety of combinations of site-specifically deuterated and/or 13C-enriched NTPs that can subsequently be used for in vitro transcription (Scott et al. 2000). Incorporation of rNTPs with selectively deuterated ribose groups led to dramatic simplification of NMR spectra obtained for a 30 KD (86 nt) GGAA tetra loop receptor complex (Fig. 3), and enabled its structure determination (Davis et al. 2005).
Reaction scheme for the enzymatic conversion of glucose to the four NTPs used to make RNA. Glucose and all intermediates are shown in boldface type, and enzymes denoted in italics. Reprinted with permission from Scott et al. (2000)
Relief of spectral crowding by site-specific deuteration. a Secondary structure of a 30 KDa GAAA tetra loop–receptor RNA. The helical regions are shown in black, the tetra loop is shown in red and the receptor is in green. b, c: portions of the 2D NOESY spectra obtained for the fully-protonated b and selectively deuterated c tetra loop receptor RNA. Reprinted, with permission, from Davis et al. (2005)
Perdeutarated NTPs
Perdeutarated NTPs are commercially available and can be used in combination with protonated NTPs to enzymatically synthesize RNAs in which specific types of nucleotides are protonated (with the remainder being partially or fully deuterated). The main advantage of this approach is that it allows spectral editing without the significant signal broadening associated with 13C incorporation. In fact, 1H NMR signals of partially deuterated samples are typically narrower than those observed for fully protonated samples, due to limited 1H–1H spin diffusion. Sequential nucleotides of a given type can be readily assigned via the standard sequential walk strategy (Wüthrich 1986). A significant disadvantage is that samples containing more than one type of protonated NTP may be required in order to make sequential signal assignments for adjacent residues of different types. Fortunately, the C8–2H deuterons can be exchanged with protons, providing a convenient means of assigning fully protonated nucleotides that are adjacent to C8-protonated, ribose-deuterated purines (Fig. 4). This approach, combined with traditional 3D and 4D 13C-edited NOESY experiments, were used to complete the NMR signal assignment and structure determination of a 101-nt viral RNA encapsidation signal (Fig. 4) (D’Souza et al. 2004). Studies of a 30-kDa GAAA tetraloop-receptor complex (Davis et al. 2005) required more sensitive two-dimensional filtered/edited NOESY experiments developed by Feigon and co-workers (Peterson et al. 2004), but even with these experiments many signals were broadened beyond detection.
Simplification of the 2D NOESY spectrum obtained for nucleotide-specific protonated, 2H-labeled RNA. a Secondary structure of the 101-nt core encapsidation signal of the Moloneymurine leukemia virus. b Portion of the 2D NOESY spectrum of the G-protonated, C,U,A-perdeuterated RNA. Breakthrough peaks from a small amount of A-H8 substitution facilitated assignment of the NMR signals. Reprinted, with permission, from D’Souza et al. (2004)
H8 enrichment of perdeuterated purines
Perdeuterated purine rNTPs that are presently available from commercial sources typically contain a low to modest levels of protonation at the C8 position (ca. 5–30%). As indicated above, specific protonation at this site can greatly facilitate assignment and analysis of 2D NOESY spectra obtained for partially deuterated RNAs, and a greater level of protonation at this site can be advantageous. It is well known that purine H8 protons readily exchange with deuterium under basic conditions (Rabi and Fox 1973; Goodman 1974). Triethylamine (TEA) is a suitable reagent for promoting 1H/2H exchange at the C8 position because of its ease of handling (Huang et al. 1997). TEA is volatile and can be removed after 1H/2H exchange simply by lyophilization. In our laboratory, commercially obtained, perdeuterated GTP and ATP are incubated with 1–5 equivalent TEA in water at 60°C for 24 h and for 5 days, respectively, which results in essentially complete exchange of D8 to H8 in both rNTPs with minimal to no rNTP hydrolysis or degradation.
2D NOESY spectra obtained for a 132 nucleotide dimeric RNA prepared with and without nucleotide-specific deuteration are shown in Fig. 5b. The fingerprint region of the fully protonated sample exhibited extreme signal overlap and was unassignable. However, well-resolved and assignable signals were observed for a sample prepared with fully perdeuterated rCTP and rUTP, H8-exchanged/perdeuterated rATP, and fully protonated rGTP (Fig. 5c). Although this approach facilitated assignment of the purine H8 and H1′ signals, assignment of the pyrimidine aromatic signals was problematic due to the large intrinsic linewidths of the H5 and H6 proton signals and the associated signal overlap.
H8 enrichment of perdeuterated purines simplifies the NMR spectrum and enables assignment of larger RNAs. a Predicted secondary structure of stem loops C and D of the Moloneymurine leukemia virus 5′-UTR. b 2D NOESY spectrum of the fully protonated SL-CD dimer. c 2D NOESY spectrum of the SL-CD dimer synthesized using fully-protonated GTP, H8-protonated, perdeuterated ATP, perdeuterated CTP and perdeuterated UTP. Black and red lines denote inter-guanosine and adenosine-to guanosine-H1′ connectivities, respectively (Miyazaki and Summers, unpublished)
H6 and H5 enrichment of perdeuterated pyrimidines
For larger RNAs, the 1H NMR signals of the H5 and H6 aromatic protons can be broadened due to 1H–1H dipolar coupling. Three-bond 1H–1H scalar coupling also contributes to the apparent linewidths of these protons, and it would be advantageous to use samples in which the C5 or C6 protons were selectively substituted by 2H. Unfortunately, procedures have not been reported for specifically substituting the H5 or H6 protons of rNTPs by deuterium. Procedures have been reported for specific C5 or C6 deuterium substitutions with pyrimidine 5′-monophosphates (Cullis et al. 1995; Rabi and Fox 1973). H5 substitution by deuterium was reportedly achieved at ~90% levels, and H6 substitution was achieved at 65–75% levels (Huang et al. 1997). In preliminary experiments, our laboratory has had difficulties achieving these levels of deuterium exchange without significant concomitant sample degradation. Preparation of selectively C6-protonated/perdeuterated rNTPs would require several steps, including hydrolysis of commercially available, perdeuterated nucleotides, 1H/2H exchange, removal of degradation contaminants, and re-phosphorylation. The field would clearly benefit by the commercial availability of perdeuterated, C6-protonated (or protonated, C5-deuterated) pyrimidine triphosphate ribonucleotide. Some of these desired rNTPs may soon be available from commercial sources.
Segmental labeling strategies
One method for overcoming signal overlap problems associated with larger RNAs involves chemically ligating a small, isotopically labeled RNA fragment to a larger, unlabeled (or differentially labeled) fragment. Segmental ligation has been most commonly performed using either T4 DNA ligase or T4 RNA ligase, each of which has advantages and disadvantages. Deoxyribozymes that catalyze RNA ligation with high efficiencies and turnover rates have also been developed recently.
T4 DNA ligase
Moore and Sharp first used T4 DNA ligase to link two RNA strands (Moore and Sharp 1992). T4 DNA ligase requires a DNA splint or cDNA template to align and hold the RNA fragments together (Fig. 6a). The RNA fragments to be ligated need to have correct termini at the junction site (3′-OH and 5′-monophosphate). Xu et al. used T4 DNA ligase to synthesize two different segmentally 15N labeled samples (Xu and Crothers 1996) corresponding to the 5′ half of Caenorhabditis elegans spliced leader RNA. They first prepared two full-length RNAs by in vitro transcription: one that was fully labeled and one that was unlabeled. RNase H was then used to cleave both RNAs into two fragments, both of which contained homogeneous ends at the cleavage site. The cleaved 5′-fragment of the unlabeled RNA was ligated to the cleaved 3′ fragment of the labeled RNA using T4 DNA ligase (yield ~18–54%). Comparison of the 2D 1H–15N HSQC spectra obtained for the uniformly labeled and segmentally labeled samples enabled the investigators to unambiguously distinguish between two competing secondary structure models.
Commonly used methods for large-scale RNA ligation. a DNA ligase mediated ligation. b RNA ligase mediated ligation. c Deoxyribozyme mediated RNA ligation. RNA ligase mediated ligation can occur with or without DNA splint templating. c is reproduced from Purtha et al. (2005) with permission
Although DNA ligase has been widely used to incorporate radioactive groups, nucleotide analogs, and crosslinking groups at specific sites (Query et al. 1994; Gozani et al. 1996; Pasman and Garcia-Blanco 1996; Maroney et al. 2000; Frilander and Steitz 2001), the ligation yield is relatively low and the reaction is slow. As such, this approach is not always ideal for preparing quantities of RNAs needed for structural analysis. In addition, the intrinsic secondary structures of the DNA splint and the RNA fragments to be ligated can interfere with hybridization, resulting in reduced ligation efficiencies. The DNA splint length can also affect the ligation yield (Kurschat et al. 2005).
T4 RNA ligase
T4 RNA ligase can also be used to ligate isotopically labeled and unlabeled RNA fragments. Like DNA ligase, T4 RNA ligase requires a 5′-monophosphate on the donor fragment and a 3′-hydroxyl group on the acceptor element. Unlike DNA ligase, RNA ligase can ligate single stranded nucleotides at the junction site. (Fig. 6b) It can work effectively both in a templated (Bain and Switzer 1992) and non-templated fashion. For templated ligation, the single-stranded RNA acceptor and donor are brought in close proximity using a DNA template (also called a splint) or tRNA (Wittenberg and Uhlenbeck 1985; Ohtsuki et al. 1996; Ohtsuki et al. 1998; Kim et al. 2002). The template is designed such that it does not base pair with residues near the penultimate nucleotides that are to be ligated (typically 2–6 residues on both the donor and acceptor strands). Ligation efficiency is reduced when the ultimate and penultimate nucleotides of the acceptor molecule are pyrimidines (Wittenberg and Uhlenbeck 1985).
Several groups have used T4 RNA ligase for non-templated ligation. Watanabe and co-workers prepared nematode mitochondrial tRNAMet constructs containing one or two isotopically labeled nucleotides. Labeling was achieved by ligating labeled nucleotides to one RNA segment, then appending a second RNA strand (Ohtsuki et al. 1996, 1998). The un-reacted substrate from the first ligation step was oxidized with periodate to avoid a purification step (Kurata et al. 2003). Based on NMR data obtained for these specifically-labeled samples, a tertiary structural model for the tRNA could be constructed (Ohtsuki et al. 1996, 1998).
Puglisi and co-workers used T4 RNA ligase to segmentally label a 100 kDa IRES RNA (Kim et al. 2002). A simple one step reaction was used to prepare the RNA fragments for ligation. For the acceptor, a hammerhead ribozyme was attached to the 5′ without the need to treat with calf intestinal alkaline phosphatase to dephosphate. For the donor, they attached a 3′-hammerhead sequence, which leaves a 2′,3′-cyclic phosphate after cleavage, and blocks the 3′ terminus from self-ligation. GMP was used to prime the transcription reaction for the donor fragment in order to obtain the appropriate 5′-monophosphate. The acceptor and donor RNAs were then ligated into a 100 kDa RNA by T4 RNA ligase, with ligation yields of up to ~50%. NMR data obtained for the ligated construct indicated that domain II of the IRES forms an independent structure that does not interact with other parts of IRES (Kim et al. 2002).
Lukavsky and co-workers developed a more convenient ligation strategy, in which the donor and acceptor RNA fragments are obtained from a single template (Lukavsky and Puglisi 2005; Tzakos et al. 2006, 2007). In the plasmid template, a T7 promoter sequence is followed by a donor sequence and a 3′-hammerhead ribozyme (Fig. 7). The 3′ ribozyme is connected to a 5′ ribozyme sequence via a short, flexible linker. The reaction is primed with GMP, and the primary transcript is excised by the HH ribozymes to yield the desired donor fragment with 5′-monophosphate and 3′-cyclic phosphate. The acceptor fragment contains hydroxyl groups on both termini. With this strategy, four ligation fragments (two enriched with 15N and two with 13C) can be obtained from only two transcription reactions. The method was used to prepare segmentally labeled samples of a 25 kDa brain cytoplasmic RNA. A potential concern with this approach is that it does not provide a means of controlling 3′-terminus run-off transcription, and non-native nucleotides could therefore be incorporated into the middle of the full length RNA.
Scheme for preparation of segmentally labeled RNAs from a single template. (a) Plasmid template, in which the 3′-hammerhead (HH) and 5′-HH were engineered into the donor and acceptor RNAs respectively. (b–d): Representative aromatic regions of 1H,13C-TROSY spectra of BC1 DTE RNA recorded with (b) a uniformly 13C-/15 N-labeled sample, (c) a 13C-SI,15N-SII segmentally labeled sample and (d) a 15N-SI,13C-SII segmentally labeled sample. Reproduced from Tzakos et al. (2007) with permission
Our group has used T4 RNA ligase to prepare the segmentally labeled HIV-1 5′-UTR. A HDV ribosome sequence was designed into the 3′-ends of the acceptor and donor fragments to control the homogeneity and prevent self-ligation. Following HDV cleavage, the acceptor 3′-cyclic phosphate was removed using T4 polynucleotide kinase. The ligation yield of using this approach (without a splint) exceeded 95%. High quality 1H,13C-correlated HMQC spectra were obtained for the 712 nucleotide dimeric 5′-UTR (Fig. 8), showing clearly that signal assignments and structural studies should be feasible for very large RNAs with appropriate internal dynamics (unpublished results).
Representative 1H–13C HMQC NMR spectrum obtained for the AUG region of the intact, 712 nt dimeric HIV-1 5′-UTR. a One of several predicted secondary structures of HIV-1NL4-3 5′-UTR; DIS, dimer initiation site; SD, major splice donor site; AUG, (green, bold), gag start codon. 13C-labeled residues are shown in green. b 1H–13C HMQC spectrum obtained for the dimeric HIV-1 5′-UTR at low ionic strength (10 mM Tris–HCl, pH 7.0) (Lu and Summers, unpublished)
Deoxyribozyme-catalyzed synthesis of 3′-5′ RNA linkages
DNA and RNA ligases do not always provide acceptable yields, and deoxyribozymes have been developed as a potentially attractive alternative for RNA ligation. Deoxyribozymes are DNA enzymes that typically catalyze reactions involving nucleic acid substrates (Höbartner and Silverman 2007). Silverman and co-workers engineered two RNA ligase deoxyribozymes that generate 3′-5′ linkages rapidly and in high yield (Fig. 6c) (Purtha et al. 2005). The deoxyribozyme has modest sequence requirements for the RNA substrates, and the 5′- end of the donor does not require to be monophosphate.
Three fragment RNA ligation
Rader and coworkers found that the DNA splint normally used for DNA ligase can significantly improve the catalytic efficiency of T4 RNA ligase-mediated RNA ligation (Stark et al. 2006). A 128 nt ligated RNA was generated from three RNA fragments using a single template with yields as high as 70–80% (5 min reaction time). All three fragments were generated by chemical synthesis, so end heterogeneity was not an issue. Also, because the DNA splint brings the appropriate 5′- and 3′- ends into close proximity, self-ligation was not a significant problem. In addition, the RNA ligase did not exhibit sequence specificity for the ligating substrates. Similar yields were obtained for a two-step ligation procedure, in which two of the fragments were ligated first, followed by ligation of the third fragment.
Wijmenga and co-workers also demonstrated that isotopically labeled uridine can be incorporated into the central position of the 20 kDa є-RNA of Duck Hepatitis B virus via three fragment RNA ligation (Nelissen et al. 2008). The optimal ligation site was chosen based on the DINAMelt (Markham and Zuker 2005; Markham and Zuker 2008) predicted tendency of the 3′- and 5′-ends of the isolated fragments to remain unstructured prior to ligation. Two protocols were evaluated: a two-step protocol that uses T4 DNA ligase and RNA ligase 1, and a one-pot protocol, that uses T4 RNA ligase 1 only. The one-pot protocol gave somewhat lower yields and more side products. This method allowed direct observation of a specific imino proton in the 20 kD aligated RNA (Nelissen et al. 2008).
Conclusions
Although the field of RNA structural biology has not kept pace with that of proteins, recent advances in nucleic acid molecular biology, coupled with the development of new NMR methodologies, should significantly facilitate future productivity. Computational tools and reagents for the design, synthesis and ligation of RNA fragments are convenient to use and cost-efficient. New approaches for assigning NMR spectra and for determining RNA structure and dynamics that have been developed over the past two decades are capable of providing mechanistic insights that cannot be obtained by other methods. The stage appears to be nearly set for answering some of the major questions in the expanding field of RNA biology.
A significant limitation to studies of larger RNAs by NMR is that the variety of commercially available, isotopically labeled rNTPs is limited, and those that can be purchased are relatively expensive. Aside from reducing the costs, the most immediate need is for pyrimidine rNTPs that are either selectively protonated at C6 and perdeuterated at all (or most) other non-exchangeable C–H sites, or selectively deuterated at C5 and protonated at all (or most) other sites. Such reagents would complement the C8-protonated purine rNTPs that can readily be generated from commercial, perdeuterated rNTPs. A second major need is for commercially available, isotopically labeled ribonucleotide phosophoramidites that could be used to chemically synthesize relatively short RNAs with labels at specific positions. Such fragments could then be enzymatically ligated with larger fragments to allow direct observation of specific nucleotides of interest under native-like (functionally active) conditions.
References
Aboul-ela F, Karn J, Varani G (1996) Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res 397:4–3981
Abramovitz DL, Pyle AM (1997) Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J Mol Biol 266:493–506
Allain FH-T, Varani G (1997) How accurately and precisely can RNA structure be determined by NMR? J Mol Biol 267:338–351
Amarasinghe GK, De Guzman RN, Turner RB, Summers MF (2000) NMR structure of stem loop SL2 of the HIV-1 Y-RNA packaging signal reveals a novel A-U-A base triple platform. J Mol Biol 299:145–156
Anderson AC, Scaringe SA, Earp BE, Frederick CA (1996) HPLC purification of RNA for crystallography and NMR. RNA 2:110–117
Azarani A, Hecker KH (2001) RNA analysis by ion-pair reversed-phase high performance liquid chromatography. Nucleic Acid Res 29:e7
Bain JD, Switzer C (1992) Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res 20:4372
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Batey RT, Kieft JS (2007) Improved native affinity purification of RNA. RNA 13:1384–1389
Batey RT, Inada M, Kujawinski E, Puglisi JD, Williamson JR (1992) Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res 20:4515–4523
Batey RT, Battiste JL, Williamson JR (1995) Preparation of isotopically enriched RNAs for heteronuclear NMR. Methods Enzymol 261:300–322
Berkower I, Leis JH J (1973) Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid-deoxyribonucleic acid hybrid structures. J Biol Chem 248:5914–5921
Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R (2008) Isolation of an active step I spliceosome and composition of its RNP core. Nature 452:846–850
Birikh KR, Heaton PA, Eckstein F (1997) The structure, function and application of the hammerhead enzyme. Eur J Biochem 245:1–16
Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI (2007) The multifunctional nucleolus. Nat Rev Mol Cell Biol 8:574–585
Brodersen P, Voinnet O (2006) The diversity of RNA silencing pathways in plants. Trends Genet 22:268–280
Cerrone-Szakal AL, Chadalavada DM, Goldend BL, Bevilacqua PC (2009) Mechanistic characterization of the HDV genomic ribozyme: The cleavage site base pair plays a structural role in facilitating catalysis. RNA 14:1746–1760
Cheong HK, Hwang E, Lee C, Choi BS, Cheong C (2004) Rapid preparation of RNA samples for NMR spectroscopy and X-ray crystallography. Nucleic Acids Res 32:e84
Crowe J, Döbeli H, Gentz R, Hochuli E, Stuber D, Henco K (1994) 6xHis-Ni–NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol Biol 31:371–387
Cullis PM, Malone ME, Podomore ID, Symons MCR (1995) Site of protonation of one-electron-reduced cytosine and its derivatives in aqueous methanol glasses. J Phys Chem 99:9293–9298
D’Souza V, Dey A, Habib D, Summers MF (2004) NMR structure of the 101 nucleotide core encapsidation signal of the Moloney Murine Leukemia Virus. J Mol Biol 337:427–442
Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE (2005) RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol 351:371–382
Dayie KT (2008) Key labeling technologies to tackle sizeable problems in RNA structural biology. Int J Mol Sci 9:1214–1240
Doudna JA, Rath VL (2002) Structure and function of the eukaryotic ribosome: the next frontier. Cell 109:153–156
Edwards TE, Klein DJ, Ferre-D’Amare AR (2007) Riboswitches: small-molecule recognition by gene regulatory RNAs. Curr Opin Struct Biol 17:273–279
Fiala R, Czernek J, Sklenar V (2000) Transverse relaxation optimized triple-resonance NMR experiments for nucleic acids. J Biomol NMR 16:291–302
Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K (2004) Uniform and residue-specific 15 N-labeling of proteins on a highly deuterated background. J Biomol NMR 29:289–297
Frilander MJ, Steitz JA (2001) Dynamic exchanges of RNA interactions leading to catalytic core formation in the U12-dependent spliceosome. Mol Cell 7:217–226
Goodman L (1974) Chemical synthesis and transformations of nucleosides. In: Ts’o PO (ed) Basic principles in nucleic acid chemistry. Academic Press, New York, pp 170–208
Gozani O, Feld R, Reed R (1996) Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10:233–243
Hartmann RK, Bindereif A, Schön A, Westhof E (2005) Hand book of RNA biochemistry. Wiley, VCH
Hassouna N, Michot B, Bachellerie JP (1984) The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res 12:3563–3583
Helm M, Brulé H, Giegé R, Florentz C (1999) More mistakes by T7 RNA polymerase at the 5’ ends of in vitro-transcribed RNAs. RNA 5:618–621
Höbartner C, Silverman SK (2007) Review: recent advances in DNA catalysis. Biopolymers. Biopolymers 87:279–291
Huang XY P, LeProust E, Gao X (1997) An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acid Res 25:4758–4763
Inoue H, Hayase Y, Iwai S, Ohtsuka E (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett 215:327–330
Kainosho M, Torizawa T, Iwashita Y, Terauchi T, Mei Ono A, Guntert P (2006) Optimal isotope labelling for NMR protein structure determinations. Nature 440:52–57
Kao C, Zheng M, Rüdisser S (1999) A simple and efficient method to reduce non templated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5:1268–1272
Kim VN (2005) Small RNAs: classification, biogenesis, and function. Mol Cells 19:1–15
Kim I et al (1995) Selective deuteration of RNA for NMR signal assignment. Nucleic Acids Symp Ser 34:123–124
Kim I, Lukavsky PJ, Puglisi JD (2002) NMR study of 100 kDa HCV IRES RNA using segmental isotope labeling. J Am Chem Soc 124:9338–9339
Kim I, McKenna SA, Viani-Puglisi E, Puglisi JD (2007) Rapid purification of RNAs using fast performance liquid chromatography (FPLC). RNA 13:289–294
Korostelev A, Noller HF (2007) The ribosome in focus: new structures bring new insights. Trends Biochem Sci 32:434–441
Kurata S, Ohtsuki T, Suzuki T, Watanabe K (2003) Quick two-step RNA ligation employing periodate oxidation. Nucl Acids Res 31:e145
Kurschat WCM J, Wombacher R, Helm M (2005) Optimizing splinted ligation of highly structured small RNAs. RNA 11:1909–1914
Lapham J, Crothers DM (1996) RNase H cleavage for processing of in vitro transcribed RNA for NMR studies and RNA ligation. RNA 2:289–296
Lukavsky PJ, Puglisi JD (2004) Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA 10:889–893
Lukavsky PJ, Puglisi JD (2005) Structure determination of large biological RNAs. Methods Enzymol 394:399–415
Lukavsky PJ, Kim I, Otto GA, Puglisi JD (2003) Structure of HCV IRES domain II determined by NMR. Nature Struct Biol 10:1033–1038
Markham NR, Zuker M (2005) DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res 33:W577–W581
Markham NR, Zuker M (2008) UNAFold: software for nucleic acid folding and hybridization. Methods Mol Biol 453:3–31
Maroney PA, Romfo CM, Nilsen TW (2000) Functional recognition of 5′ splice site by U4/U6.U5 tri-snRNP defines a novel ATP-dependent step in early spliceosome assembly. Mol Cell 6:317–328
McKenna SA et al (2007) Purification and characterization of transcribed RNAs using gel filtration chromatography. Nat Protoc 2:3270–3277
Michnicka MJ, Harper JW, King GC (1993) Selective isotopic enrichment of synthetic RNA: application to the HIV-1 TAR element. Biochemistry 32:395–400
Milligan JF, Uhlenbeck OC (1989) Synthesis of small RNAs using T7 RNA polymerase. Meth Enzymol 180:51–62
Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res 15:8783–9798
Moore MJ, Sharp PA (1992) Site-specific modification of pre-mRNA: the 2’-hydroxyl groups at the splice sites. Science 256:992–997
Nelissen FHT, van Gammeren AJ, Tessari M, Girard FC, Heus HA, Wijmenga SS (2008) Multiple segmental and selective isotope labeling of large RNA for NMR structural studies. Nucl Acids Res 36:e89
Nikonowicz EP, Pardi A (1992) Three-dimensional heteronuclear NMR studies of RNA. Nature 355:184–186
Ohtsuki T, Kawai G, Watanabe Y, Kita K, Nishikawa K, Watanabe K (1996) Preparation of biologically active Ascaris suum mitochondrial tRNAMet with a TV-replacement loop by ligation of chemically synthesized RNA fragments. Nucl Acids Res 24:662–667
Ohtsuki T, Kawai G, Watanabe K (1998) Stable isotope-edited NMR analysis of Ascaris suum mitochondrial tRNAMet having a TV-replacement loop. J Biochem 124:28–34
Pasman Z, Garcia-Blanco MA (1996) The 5′ and 3′ splice sites come together via a three dimensional diffusion mechanism. Nucleic Acids Res 24:1638–1645
Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Peterson RD, Theimer CA, Wu H, Feigon J (2004) New applications of 2D filtered/edited NOESY for assignment and structure elucidation of RNA and RNA–protein complexes. J Biomol NMR 28:59–67
Pleiss JA, Derrick ML, Uhlenbeck OC (1998) T7 RNA polymerase produces 5’ end heterogeneity during in vitro transcription from certain templates. RNA 4:1313–1317
Pokrovskaya ID, Gurevich VV (1994) In vitro transcription: preparative RNA yields in analytical scale reactions. Anal Biochem 220:420–423
Ponting CP, Oliver PL, Reik W (2009) Evolution and functions of long no ncoding RNAs. Cell 136:629–641
Purtha WE, Coppins RL, Smalley MK, Silverman SK (2005) General deoxyribozyme-catalyzed synthesis of native 3′-5′ RNA linkages. J Am Chem Soc 127:13124–13125
Query CC, Moore MJ, Sharp PA (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev 8:587–597
Rabi JA, Fox JJ (1973) Nucleosides. LXXIX. Facile base-catalyzed hydrogen isotope labeling at position 6 of pyrimidine nucleosides. J Am Chem Soc 95:1628–1632
Riek R, Wider G, Pervushin K, Wuthrich K (1999) Polarization transfer by cross-correlated relaxation in solution NMR with very large molecules. Proc Natl Acad Sci USA 96:4918–4923
Scaringe SA, Wincott FE, Caruther MH (1998) Novel RNA synthesis method using 5′-O-Silyl-2′-O-orthoester protecting groups. J Am Chem Soc 120:11820–11821
Scott LG, Tolbert TJ, Williamson JR (2000) Preparation of specifically 2H- and 13C-labeled ribonucleotides. Methods Enzymol 317:18–38
Shah SA, Brunger AT (1999) The 1.8 Å crystal structure of a statically disordered 17 base pair RNA duplex: Principles of RNA crystal packing and its effect on nucleic acid structure. J Mol Biol 285:1577–1588
Shields TP, Mollova E, Ste marie L, Hansen MR, Pardi A (1999) High-performance liquid chromatography purification of homogeneous-length RNA produced by trans cleavage with a hammerhead ribozyme. RNA 5:1259–1267
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20s proteasome by NMR. Nature 445:618–622
Spriggs S, Garyu L, Connor R, Summers MF (2008) Potential intra- and intermolecular interactions involving the Unique-5′ Region of the HIV-1 5′-UTR. Biochemistry 46:13064–13073
Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD (2006) An RNA ligase-mediated method for the efficient creation of large, synthetic RNAs. RNA 12:2014–2019
Steitz TA (2008) A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9:242–253
Tzakos AG, Grace CRR, Lukavsky PJ, Riek R (2006) NMR techniques for very large proteins and RNAs in solution. Annu Rev Biophys Biomol Str 35:319–342
Tzakos AG, Easton LE, Lukavsky PJ (2007) Preparation of large RNA oligonucleotides with complementary isotope-labeled segments for NMR structural studies. Nat Protoc 2:2139–2147
Usman N, Ogilvie KK, Jiang MY, Cedergren RJ (1987) The automated chemical synthesis of long oligoribuncleotides using 2′-O-silylated ribonucleoside 3′-O-phosphoramidites on a controlled-pore glass support: synthesis of a 43-nucleotide sequence similar to the 3’-half molecule of an Escherichia coli formylmethionine tRNA. J Am Chem Soc 109:7845–7854
Wakeman CA, Winkler WC, Dann CE III (2007) Structural features of metabolite-sensing ribo switches. Trends Biochem Sci 32:415–424
Wittenberg WL, Uhlenbeck OC (1985) Specific replacement of functional groups of uridine-33 in yeast phenylalanine transfer ribonucleic acid. Biochemistry 24:2705–2712
Wüthrich K (1986) NMR of proteins and nucleic acids. Wiley, New York
Wyatt JR, Chastain M, Puglisi JD (1991) Synthesis and purification of large amounts of RNA oligonucleotides. BioTechniques 11:764–769
Xu JL J, Crothers DM (1996) Determining RNA solution structure by segmental isotopic labeling and NMR: application to Caenorhabditis elegans spliced leader RNA 1. Proc Natl Acad Sci USA 93:44–48
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Lu, K., Miyazaki, Y. & Summers, M.F. Isotope labeling strategies for NMR studies of RNA. J Biomol NMR 46, 113–125 (2010). https://doi.org/10.1007/s10858-009-9375-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-009-9375-2