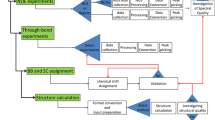
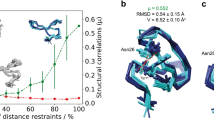
Abstract
Protein structure determination by NMR methods has started in the mid-eighties and has been growing steadily since then. Ca. 14% of the protein structures deposited in the PDB have been solved by NMR. The evaluation of the quality of NMR structures however is still lacking a well-established practice. In this work, we examined various tools for the assessment of structural quality to ascertain the extent to which these tools could be applied to detect flaws in NMR structures. In particular, we investigated the variation in the scores assigned by these programs as a function of the deviation of the structures induced by errors in assignments or in the upper distance limits used. These perturbations did not distort radically the protein fold, but resulted in backbone RMS deviations up to 3 Å, which is in line with errors highlighted in the available literature. We found that it is quite difficult to discriminate the structures perturbed because of misassignments from the original ones, also because the spread in score over the conformers of the original bundle is relatively large. ϕ–ψ distributions and normality scores related to the backbone conformation and to the distribution of side-chain dihedral angles are the most sensitive indicators of flaws.


Similar content being viewed by others
Abbreviations
- NMR:
-
Nuclear magnetic resonance
- NOE:
-
Nuclear Overhauser effect
- RMSD:
-
Root mean standard deviation
References
Andrec M, Snyder DA, Zhou Z, Young J, Montelione GT, Levy RM (2007) A large data set comparison of protein structures determined by crystallography and NMR: statistical test for structural differences and the effect of crystal packing. Proteins 69:449–465
Banci L, Bertini I Cantini F, Migliardi M, Rosato A, Wang S (2005) An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A. J Mol Biol 352:409–417
Barbieri R, Luchinat C, Parigi G (2004) Backbone-only protein solution structures with a combination of classical and paramagnetism-based constraints: a method that can be scaled to large molecules. ChemPhysChem 5(6):797–806
Bhattacharya A, Tejero R, Montelione GT (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66:778–795
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucl Acids Res 28:235–242
Bertini I, Cavallaro G, Luchinat C, Poli I (2003) A use of Ramachandran potentials in protein solution structure determinations. J Biomol NMR 4:355–366
Bertini I, Donaire A, Jimenez B, Luchinat C, Parigi G, Piccioli M, Poggi L (2001) Paramagnetism-based versus classical constraints: an analysis of the solution structure of Ca Ln calbindin D9k. J Biomol NMR 21:85–98
Billeter M (1992) Comparison of protein structures determined by NMR in solution and by X-ray diffraction in single crystals. Q Rev Biophys 25:325–377
Branden CI, Jones TA (1990) Between objectivity and subjectivity. Nature 343:687–689
Brown EN, Ramaswamy S (2007) Quality of protein crystal structures. Acta Crystallogr D Biol Crystallogr 63:941–950
Brünger AT, Clore GM, Gronenborn AM, Saffrich R, Nilges M (1993) Assessing the quality of solution nuclear magnetic resonance structures by complete cross-validation. Science 261:328–331
Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA (2004) AMBER 8. University of California, San Francisco
Clore GM, Robien MA, Gronenborn AM (1993) Exploring the limits of precision and accuracy of protein structures determined by nuclear magnetic resonance spectroscopy. J Mol Biol 231:82–102
Cornilescu G, Marquardt JL, Ottiger M, Bax A (1999) Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. J Am Chem Soc 120:6836–6837
Davis IW, Murray LW, Richardson JS, Richardson DC (2007) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32(Web Server issue):W615–W619
DePristo MA, de Bakker PI, Blundell TL (2004) Heterogeneity and inaccuracy in protein structures solved by X-ray crystallography. Structure 12:831–838
Doreleijers JF, Rullmann JA, Kaptein R (1998) Quality assessment of NMR structures: a statistical survey. J Mol Biol 281:149–164
Doreleijers JF, Mading S, Maziuk D, Sojourner K, Yin L, Zhu J, Markley JL, Ulrich EL (2003) BioMagResBank database with sets of experimental NMR constraints corresponding to the structures of over 1400 biomolecules deposited in the Protein Data Bank. J Biomol NMR 26:139–146
Duquesne AE, de Ruijter M, Brouwer J, Drijfhout JW, Nabuurs SB, Spronk CAEM, Vuister GW, Ubbink M, Canters GW (2005) Solution structure of the second PDZ domain of the neuronal adaptor X11alpha and its interaction with the C-terminal peptide of the human copper chaperone for superoxide dismutase. J Biomol NMR 32:209–218
Garbuzynskiy SO, Melnik BS, Lobanov MY, Finkelstein AV, Galzitskaya OV (2005) Comparison of X-ray and NMR structures: is there a systematic difference in residue contacts between X-ray- and NMR-resolved protein structures? Proteins 60:139–147
Gronwald W, Moussa S, Elsner R, Jung A, Ganslmeier B, Trenner J, Kremer W, Neidig KP, Kalbitzer HR (2002) Automated assignment of NOESY NMR spectra using a knowledge based method (KNOWNOE). J Biomol NMR 23:271–287
Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Bio 273:283–298
Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 24:209–227
Huang YJ, Moseley HN, Baran MC, Arrowsmith C, Powers R, Tejero R, Szyperski T, Montelione G (2005a) An integrated platform for automated analysis of protein NMR structures. Methods Enzymol 394:111–141
Huang YJ, Powers R, Montelione GT (2005b) Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J Am Chem Soc 127:1665–1674
Hooft RWW, Vriend G, Sander C, Abola EE (1996a) Errors in protein structures. Nature 381:272
Hooft RW, Sander C, Vriend G (1996b) Verification of protein structures: side-chain planarity. J Appl Cryst 29:714–716
Hooft RWW, Sander C, Vriend G (1997) Objectively judging the quality of a protein structure from a Ramachandran plot. Comput Appl Biosci 13:425–430
Joosten RP, Vriend G (2007) PDB improvement starts with data deposition. Science 317:195–196
Kleywegt GJ (2000) Validation of proteins crystal structures. Acta Crystallogr D 56:249–265
Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structure. J Appl Cryst 26:283–291
Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486
Lovell SC, Davis IW, Arendall WB III, de Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50:437–450
Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J Biomol NMR 12:1–23
Morris AL, MacArthur MW, Hutchinson EG, Thornton JM (1992) Stereochemical quality of protein structures coordinates. Proteins 12:345:364
Moseley HN, Sahota G, Montelione GT (2004) Assignment validation software suite for the evaluation and presentation of protein resonance assignment data. J Biomol NMR 28:341–355
Nabuurs SB, Krieger E, Spronk CA, Nederveen AJ, Vriend G, Vuister GW (2005) Definition of a new information-based per-residue quality parameter. J Biomol NMR 33:123–134
Nabuurs SB, Nederveen AJ, Vranken W, Doreleijers JF, Bonvin AM, Vuister GW, Vriend G, Spronk CA (2004) DRESS: a Database of Refined Solution NMR Structures. Proteins 55:483–486
Nabuurs SB, Spronk CA, Vuister GW, Vriend G (2006) Traditional biomolecular structure determination by NMR spectroscopy allows for major errors. PLoS Comput Biol 2(2):e9. doi:10.1371/journal.pcbi.0020009
Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM (2005) RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59:662–672
Pugalenthi G, Shameer K, Srinvasan N, Sowdhamini R (2006) HARMONY: a server for the assessment of protein structures. Nucl Acids Res 34(Web Server issue):W231–W234
Ramachandran GN, Ramakrishnan C, Sasisekharan V (1963) Stereochemistry of polypeptide chain configuration. J Mol Biol 7:95–99
Ramage R, Green J, Muir TW, Ogunjobi OM, Love S, Shaw K (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J 299:151–158
Samudrala R, Moult J (1998) An all-atom distance-dependent conditional probability discriminatory function for protein structure prediction. J Mol Biol 275:895–916
Sims GE, Kim S-H (2006) A method for evaluating the structural quality of protein models by using higher-order φ–ψ pairs scoring. Proc Natl Acad Sci USA 103(12):4428–4432
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17:355–362
Spronk CA, Linge JP, Hilbers CW, Vuister GW (2002) Improving the quality of protein structures derived by NMR spectroscopy. J Biomol NMR 22:281–289
Spronk CA, Nabuurs SB, Bonvin AM, Krieger E, Vuister GW, Vriend G (2003) The precision of NMR structure ensembles revisited. J Biomol NMR 25:225–234
Tosatto SCE (2005) The Victor/FRST function for model quality estimation. J Comput Biol 12:1326–1327
Tosatto SCE, Battistutta R (2007) TAP score: torsion angle propensity normalization applied to local protein structure evaluation. BMC Bioinformatics 8:155–168
van Rossum G, Drake FL (eds) (2001) Python reference manual. PythonLabs, Virginia. Available via www.python.org
Vriend G, Sander C (1993) Quality controls of protein models: directional atomic contact analysis. J Appl Cryst 26:47–60
Wagner G, Hyberts SG, Havel TF (1992) NMR structure determination in solution: a critique and comparison with X-ray crystallography. Annu Rev Biophys Biomol Struct 21:167–198
Wanker WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas ML, Eldon JL, Ulrich MJI, Ionides J, Laue ED (2005) The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59:687–696
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35(Web Server issue):W407–W410
Acknowledgements
This work was stimulated by the activities of the Coordination Action “NMR-Life” (funded by the European Commission, project no. 18758). We thank Prof. Ivano Bertini for many useful discussions. We thank the Ente Cassa di Risparmio di Firenze, the EC (project no. 213010), and MIUR (project PRIN 2005) for financial support. E.S. is the recipient of a fellowship from the FiorGen Foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Saccenti, E., Rosato, A. The war of tools: how can NMR spectroscopists detect errors in their structures?. J Biomol NMR 40, 251–261 (2008). https://doi.org/10.1007/s10858-008-9228-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-008-9228-4