Abstract
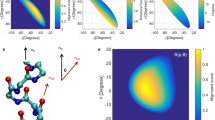
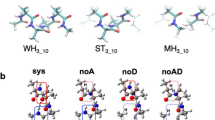
The dependence of the 13C chemical shift on side-chain orientation was investigated at the density functional level for a two-strand antiparallel β-sheet model peptide represented by the amino acid sequence Ac-(Ala)3-X-(Ala)12-NH2 where X represents any of the 17 naturally occurring amino acids, i.e., not including alanine, glycine and proline. The dihedral angles adopted for the backbone were taken from, and fixed at, observed experimental values of an antiparallel β-sheet. We carried out a cluster analysis of the ensembles of conformations generated by considering the side-chain dihedral angles for each residue X as variables, and use them to compute the 13C chemical shifts at the density functional theory level. It is shown that the adoption of the locally-dense basis set approach for the quantum chemical calculations enabled us to reduce the length of the chemical-shift calculations while maintaining good accuracy of the results. For the 17 naturally occurring amino acids in an antiparallel β-sheet, there is (i) good agreement between computed and observed 13Cα and 13Cβ chemical shifts, with correlation coefficients of 0.95 and 0.99, respectively; (ii) significant variability of the computed 13Cα and 13Cβ chemical shifts as a function of χ1 for all amino acid residues except Ser; and (iii) a smaller, although significant, dependence of the computed 13Cα chemical shifts on χξ (with ξ ≥ 2) compared to χ1 for eleven out of seventeen residues. Our results suggest that predicted 13Cα and 13Cβ chemical shifts, based only on backbone (φ,ψ) dihedral angles from high-resolution X-ray structure data or from NMR-derived models, may differ significantly from those observed in solution if the dihedral-angle preferences for the side chains are not taken into account.






Similar content being viewed by others
References
Burgess AW, Ponnuswamy PK, Scheraga HA (1974) Israel J Chem 12:239–286
Chakrabarti P, Pal D (1998) Protein Eng 11:631–647
Chesnut DB, Moore KD (1989) J Comp Chem 10:648–659
Chothia C, Levitt M, Richardson D (1977) Proc Natl Acad Sci USA 74:4130–4134
Chothia C, Janin J (1981) Proc Natl Acad Sci USA 78:4146–4150
Chou K-C, Scheraga HA (1982) Proc Natl Acad Sci USA 79:7047–7051
Chou K-C, Pottle M, Némethy G, Ueda Y, Scheraga HA (1982) J Mol Biol 162:89–112
Chou PY, Fasman GD (1974) Biochemistry 13:211–222
Creighton TE (1984) Proteins: Structure and Molecular Properties. W.E. Freeman and Company, New York, pp. 186, 223
Derrick JP, Wigley DB (1994) J Mol Biol 243:906–918
Dumbrack RL Jr, Karplus M (1993) J Mol Biol 230:543–574
Dumbrack RL Jr, Karplus M (1994) Nat Struct Biol 1:334–340
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA (1998) Gaussian 98, Revision A.7, Inc., Pittsburgh, PA
Havlin RH, Le H, Laws DD, de Dios AC, Oldfield E (1997) J Am Chem Soc 119:11951–11958
Hehre WJ, Radom L, Schleyer P, Pople JA (1986) Ab Initio Molecular Orbital Theory. John Wiley and Sons, New York
Iwadate M, Asakura T, Williamson MP (1999) J Biomol NMR 13:199–211
Kruskal JB Jr (1956) Proc American Math Soc 7:48–50
Kuszewski J, Qin JA, Gronenborn AM, Clore GM (1995) J Magn Reson Ser B 106:92–96
Laws DD, Le H, de Dios AC, Havlin RH, Oldfield E (1995) J Am Chem Soc 117:9542–9546
McGregor JM, Islam SA, Sternberg MJE (1987) J Mol Biol 198:295–310
Némethy G, Gibson KD, Palmer KA, Yoon CN, Paterlini G, Zagari A, Rumsey S, Scheraga HA (1992) J Phys Chem 96:6472–6484
Pauling LC, Corey RB (1951) Proc Natl Acad Sci USA 37:729–740
Pauling LC, Corey RB (1953) Proc R Soc London Ser B 141:21–33
Pearson JG, Le H, Sanders LK, Godbout N, Havlin RH, Oldfield E (1997) J Am Chem Soc 119:11941–11950
Ripoll DR, Scheraga HA (1988) Biopolymers 27:1283–1303
Ripoll DR, Vila JA, Scheraga HA (2005) Proc Natl Acad Sci USA 102:7559–7564
Rossmeisl J, Nørskov JK, Jacobsen KW (2004) J Am Chem Soc 126:13140–13143
Santiveri CM, Rico M, Jiménez MA (2001) J Biom NMR 19:331–345
Sibanda BL, Thornton JM (1991) Methods Enzymol 202:59–82
Sosa CP, Ochterski J, Carpenter J, Frisch MJ (1998) J Comp Chem 19:1053–1063
Spera S, Bax A (1991) J Am Chem Soc 113:5490–5492
Ulmer TS, Ramirez BE, Delaglio F, Bax A (2003) J Am ChemSoc 125:9179–9191
Vila JA, Ripoll DR, Baldoni HA, Scheraga HA (2002) J Biomol NMR 24:245–262
Vila JA, Baldoni HA, Ripoll DR, Scheraga HA (2003) J Biomol NMR 26:113–130
Vila JA, Baldoni HA, Scheraga HA (2004a) Protein Sci 13:2939–2948
Vila JA, Baldoni HA, Scheraga HA (2004b) Proteins: Struct Funct Bioinf 57:87–98
Wang Y, Jardetzky O (2002) Protein Sci 11:852–861
Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson JH, Oldfield E, Markley JL, Sykes BD. (1995) J Biomol NMR 6:135–140
Xu X-P, Case DA (2001) J Biomol NMR 21:321–333
Xu X-P, Case DA (2002) Biopolymers 65:408–423
Zhang C, Kim S-H (2000) J Mol Biol 299:1075–1089
Acknowledgments
This research was supported by grants from the National Institutes of Health (GM-14312 and TW-6335), and the National Science Foundation (MCB00-03722). Support was also received from the National Research Council of Argentina (CONICET) [PIP-02485] and from the Universidad Nacional de San Luis [UNSL] (P-328402), Argentina. This research was conducted using the resources of two Beowulf-type clusters located at (a) the Instituto de Matemática Aplicada San Luis (CONICET-UNSL) and (b) the Baker Laboratory of Chemistry and Chemical Biology, Cornell University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Villegas, M.E., Vila, J.A. & Scheraga, H.A. Effects of side-chain orientation on the 13C chemical shifts of antiparallel β-sheet model peptides. J Biomol NMR 37, 137–146 (2007). https://doi.org/10.1007/s10858-006-9118-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-006-9118-6