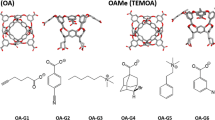
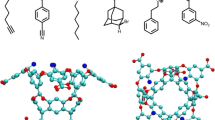
Abstract
We calculate the absolute binding free energies of tetra-methylated octa-acids host–guest systems as a part of the SAMPL6 blind challenge (receipt ID vq30p). We employed two different free energy simulation methods, i.e., the umbrella sampling (US) and double decoupling method (DDM). The US method was used with the weighted histogram analysis method (WHAM) (US-WHAM scheme). In the DDM scheme, Hamiltonian replica-exchange method (HREM) was combined with the Bennett acceptance ratio (BAR) (HREM-BAR scheme). We obtained initial binding poses via molecular docking using GalaxyDock-HG program, which is developed for the SAMPL challenge. The root mean square deviation (RMSD) and the mean absolute deviations (MAD) using US-WHAM scheme were 1.33 and 1.02 kcal/mol, respectively. The MAD was the top among all submissions, however the correlation with respect to experiment was unexceptional. While the RMSD and MAD via HREM-BAR scheme were greater than US-WHAM scheme, (i.e., 2.09 and 1.76 kcal/mol), their correlations were slightly better than US-WHAM. The correlation between the two methods was high. Further discussion on the DDM method can be found in a companion paper by Han et al. (receipt ID 3z83m) in the same issue.









Similar content being viewed by others
References
Jorgensen WL (2004) The many roles of computation in drug discovery. Science 303(5665):1813–1818
De Vivo M, Masetti M, Bottegoni G, Cavalli A (2016) Role of molecular dynamics and related methods in drug discovery. J Med Chem 59(9):4035–4061
Guthrie JP (2009) A blind challenge for computational solvation free energies: introduction and overview. J Phys Chem 113:4501–4507
Gallicchio E, Chen H, Chen H et al (2015) BEDAM binding free energy predictions for the SAMPL4 octa-acid host challenge. J Comput Aided Mol Des 29(4):315–325
Ellingson BA, Geballe MT, Wlodek S, Bayly CI, Skillman AG, Nicholls A (2014) Efficient calculation of SAMPL4 hydration free energies using OMEGA, SZYBKI, QUACPAC, and Zap TK. J Comput Aided Mol Des 28(3):289–298
Nicholls A, Mobley DL, Guthrie JP et al (2008) Predicting small-molecule solvation free energies: an informal blind test for computational chemistry. J Med Chem 51(4):769–779
Beckstein O, Fourrier A, Iorga BI (2014) Prediction of hydration free energies for the SAMPL4 diverse set of compounds using molecular dynamics simulations with the OPLS-AA force field. J Comput Aided Mol Des 28(3):265–276
Geballe MT, Skillman AG, Nicholls A, Guthrie JP, Taylor PJ (2010) The SAMPL2 blind prediction challenge: introduction and overview. J Comput Aided Mol Des 24(4):259–279
Tofoleanu F, Lee J, Pickard IVFC et al (2017) Absolute binding free energies for octa-acids and guests in SAMPL5. J Comput Aided Mol Des 31(1):107–118
Lee J, Tofoleanu F, Pickard FC et al (2017) Absolute binding free energy calculations of CBClip host–guest systems in the SAMPL5 blind challenge. J Comput Aided Mol Des 31(1):71–85
König G, Brooks BR (2012) Predicting binding affinities of host–guest systems in the SAMPL3 blind challenge: the performance of relative free energy calculations. J Comput Aided Mol Des 26(5):543–550
Gan H, Benjamin CJ, Gibb BC (2011) Nonmonotonic assembly of a deep-cavity cavitand. J Am Chem Soc 133(13):4770–4773
Gibb CLD, Gibb BC (2014) Binding of cyclic carboxylates to octa-acid deep-cavity cavitand. J Comput Aided Mol Des 28(4):319–325
Muddana HS, Fenley AT, Mobley DL, Gilson MK (2014) The SAMPL4 host–guest blind prediction challenge: an overview. J Comput Aided Mol Des 28(4):305–317
Sullivan MR, Sokkalingam P, Nguyen T, Donahue JP, Gibb BC (2017) Binding of carboxylate and trimethylammonium salts to octa-acid and TEMOA deep-cavity cavitands. J Comput Aided Mol Des 31(1):21–28
Woo H-J, Roux B (2005) Calculation of absolute protein–ligand binding free energy from computer simulations. Proc Natl Acad Sci 102(19):6825–6830
Gumbart JC, Roux B, Chipot C (2013) Efficient determination of protein–protein standard binding free energies from first principles. J Chem Theory Comput 9(8):3789–3798
Roux B (1995) The calculation of the potential of mean force using computer simulations. Comput Phys Commun 91(1–3):275–282. https://doi.org/10.1016/0010-4655(95)00053-I
Wu X, Damjanovic A, Brooks BR (2012) Efficient and unbiased sampling of biomolecular systems in the canonical ensemble: a review of self-guided Langevin dynamics. Adv Chem Phys 150:255
Wu X, Brooks BR (2003) Self-guided Langevin dynamics simulation method. Chem Phys Lett 381(3–4):512–518
Wu X, Brooks BR, Vanden-Eijnden E (2016) Self-guided Langevin dynamics via generalized Langevin equation. J Comput Chem 37(6):595–601
Wu X, Brooks BR (2011) Toward canonical ensemble distribution from self-guided Langevin dynamics simulation. J Chem Phys 134(13):04B605
Murata K, Sugita Y, Okamoto Y (2004) Free energy calculations for DNA base stacking by replica-exchange umbrella sampling. Chem Phys Lett 385(1–2):1–7
Sugita Y, Kitao A, Okamoto Y (2000) Multidimensional replica-exchange method for free-energy calculations. J Chem Phys 113(15):6042–6051
Han K, Hudson PS, Jones MR, Nishikawa N, Tofoleanu F, Brooks BR (2018) Prediction of CB [8] host–guest binding free energies in SAMPL6 using the double-decoupling method. J Comput Aided Mol Des. https://doi.org/10.1007/s10822-018-0144-8
Shin W-H, Heo L, Lee J, Ko J, Seok C, Lee J (2011) LigDockCSA: protein–ligand docking using conformational space annealing. J Comput Chem 32(15):3226–3232
Lee J, Scheraga HA, Rackovsky S (1997) New optimization method for conformational energy calculations on polypeptides: conformational space annealing. J Comput Chem 18(9):1222–1232
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28(6):1145–1152
Shin W-H, Lee GR, Seok C (2015) Evaluation of galaxydock based on the community structure–activity resource 2013 and 2014 benchmark studies. J Chem Inf Model 56(6):988–995
Shin W-H, Kim J-K, Kim D-S, Seok C (2013) GalaxyDock2: protein–ligand docking using beta-complex and global optimization. J Comput Chem 34(30):2647–2656
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31(4):671–690
Kirkwood JG (1935) Statistical mechanics of fluid mixtures. J Chem Phys 3(5):300–313
MacKerell AD Jr, Bashford D, Bellott M et al (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102(18):3586–3616
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79(2):926–935
Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4(2):187–217
Hoover WG (1985) Canonical dynamics: equilibrium phase-space distributions. Phys Rev A 31(3):1695
Nosé S (1984) A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys 81(1):511–519
Feller SE, Zhang Y, Pastor RW, Brooks BR (1995) Constant pressure molecular dynamics simulation: the Langevin piston method. J Chem Phys 103(11):4613–4621
Ryckaert J-P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23(3):327–341
Shirts MR, Chodera JD (2008) Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys 129(12):124105. https://doi.org/10.1063/1.2978177
Grossfield A. WHAM: the weighted histogram analysis method, version 2.0.9.1, http://membrane.urmc.rochester.edu/content/wham
Boresch S, Tettinger F, Leitgeb M, Karplus M (2003) Absolute binding free energies: a quantitative approach for their calculation. J Phys Chem B 107(35):9535–9551
Mobley DL, Graves AP, Chodera JD, McReynolds AC, Shoichet BK, Dill KA (2007) Predicting absolute ligand binding free energies to a simple model site. J Mol Biol 371(4):1118–1134
Gilson MK, Given JA, Bush BL, McCammon JA (1997) The statistical-thermodynamic basis for computation of binding affinities: a critical review. Biophys J 72(3):1047–1069
Itoh SG, Okumura H, Okamoto Y (2010) Replica-exchange method in van der Waals radius space: overcoming steric restrictions for biomolecules. J Chem Phys 132(13):134105
Itoh SG, Okumura H (2013) Hamiltonian replica-permutation method and its applications to an alanine dipeptide and amyloid-β (29–42) peptides. J Comput Chem 34(29):2493–2497
Fukunishi H, Watanabe O, Takada S (2002) On the Hamiltonian replica exchange method for efficient sampling of biomolecular systems: application to protein structure prediction. J Chem Phys 116(20):9058–9067
Bennett CH (1976) Efficient estimation of free energy differences from Monte Carlo data. J Comput Phys 22(2):245–268
Shirts MR, Bair E, Hooker G, Pande VS (2003) Equilibrium free energies from nonequilibrium measurements using maximum-likelihood methods. Phys Rev Lett 91(14):140601
König G, Pickard FC, Mei Y, Brooks BR (2014) Predicting hydration free energies with a hybrid QM/MM approach: an evaluation of implicit and explicit solvation models in SAMPL4. J Comput Aided Mol Des 28(3):245–257
König G, Hudson PS, Boresch S, Woodcock HL (2014) Multiscale free energy simulations: an efficient method for connecting classical MD simulations to QM or QM/MM free energies using non-Boltzmann Bennett reweighting schemes. J Chem Theory Comput 10(4):1406–1419
König G, Bruckner S, Boresch S (2009) Unorthodox uses of Bennett’s acceptance ratio method. J Comput Chem 30(11):1712–1718
Acknowledgements
The authors would like to thank Gerhard König, John Legato, Minkyung Baek and Chaok Seok for helpful discussion and technical assistance. This work was partially supported by the intramural research program of the National Heart, Lung and Blood Institute (NHLBI) of the National Institutes of Health and employed the high-performance computational capabilities of the LoBoS and Biowulf Linux clusters at the National Institutes of Health. (http://www.lobos.nih.gov and http://www.biowulf.nih.gov.)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nishikawa, N., Han, K., Wu, X. et al. Comparison of the umbrella sampling and the double decoupling method in binding free energy predictions for SAMPL6 octa-acid host–guest challenges. J Comput Aided Mol Des 32, 1075–1086 (2018). https://doi.org/10.1007/s10822-018-0166-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0166-2