Abstract
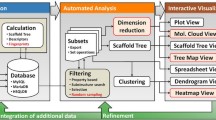
To help tracking all molecules made in a typical medicinal chemistry project, we have developed an algorithm to generate a maximum common framework (MCF) hierarchy and an interactive tool for its visualization and analysis. By identifying all unique frameworks for a set of molecules and all molecules containing each framework, we were able to simplify the MCF hierarchy build up steps and, as a result, speed up the entire process significantly. By allowing compounds to be assigned to multiple MCFs, users can easily remove bad branching nodes and concentrate on interesting ones. MCF hierarchies provide an effective and intuitive visualization for tracking medicinal chemistry lead optimization projects. We will provide examples to illustrate its usefulness.








Similar content being viewed by others
References
Pargellis C, Tong L, Churchill L, Cirillo PF, Gilmore T, Graham AG, Grob PM, Hickey ER, Moss N, Pav S, Regan J (2002) Nature Struct Biol 9:268–272
Bemis GW, Murcko MA (1996) J Med Chem 39:2887–2893
Wilkens SJ, Janes J, Su AI, Hier S (2005) J Med Chem 48:3182–3193
Schuffenhauer A, Ertl P, Roggo S, Wetzel S, Marcus AK, Waldmann H (2007) J Chem Inf Model 47:47–58
Schuffenhauer A, Brown N, Ertl P, Jenkins JL, Selzer P, Hamon J (2007) J Chem Inf Model 47:325–336
Koch MA, Schuffenhauer A, Scheck M, Wetzel S, Casaulta M, Odermatt A, Ertl P, Waldmann H (2005) PNAS 102:17272–17277
The OEChem toolkit is available from OpenEye Scientific Software, Inc. http://www.eyesopen.com
The Qt class library and tools are available from Trolltech Inc. http://www.trolltech.com
Cho SJ, Sun Y, Harte W (2006) J Comput-Aided Mol Des 20:249–261
Target Inhibitor Databases are available from GVK Biosciences Private Limited. http://www.gvkbio.com
Molecular Design Drug Data Report, version 2005.2. Molecular Design Ltd., San Leandro, CA
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cho, S.J., Sun, Y. Visual exploration of structure–activity relationship using maximum common framework. J Comput Aided Mol Des 22, 571–578 (2008). https://doi.org/10.1007/s10822-008-9206-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-008-9206-7