Abstract
Purpose
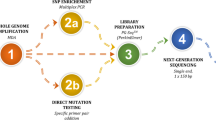
This paper aims to investigate the feasibility of performing pre-implantation genetic diagnosis (PGD) and pre-implantation genetic screening (PGS) simultaneously by a universal strategy without the requirement of genotyping relevant affected family members or lengthy preliminary work on linkage analysis.
Methods
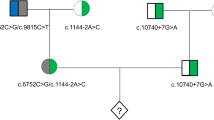
By utilizing a universal Mutated Allele Revealed by Sequencing with Aneuploidy and Linkage Analyses (MARSALA) strategy based on low depth whole genome sequencing (~3x), not involving specific primers' design nor the enrichment of SNP markers for haplotype construction. Single-sperm cells and trephectoderm cells from in vitro fertilized embryos from a couple carrying HBB mutations were genotyped. Haplotypes of paternal alleles were constructed and investigated in embryos, and the chromosome copy number profiles were simultaneously analyzed.
Results
The universal MARSALA strategy allows the selection of a euploid embryo free of disease mutations for in uterus transfer and successful pregnancy. A follow-up amniocentesis was performed at 17 weeks of gestation to confirm the PGD/PGS results.
Conclusion
We present the first successful PGD procedure based on genotyping multiple single-sperm cells to obtain SNP linkage information. Our improved PGD/PGS procedure does not require genotyping the proband or relevant family members and therefore can be applicable to a wider population of patients when conducting PGD for monogenic disorders.




Similar content being viewed by others
References
Genes and human disease: Human Genetic Programmes. 2015. http://www.who.int/genomics/public/geneticdiseases/en/index2.html
Peltonen L, Perola M, Naukkarinen J, Palotie A. Lessons from studying monogenic disease for common disease. Hum Mol Genet. 2006;15(suppl 1):R67–74.
Handyside AH, Pattinson JK, Penketh RJ, Delhanty JD, Winston RM, Tuddenham EG. Biopsy of human preimplantation embryos and sexing by DNA amplification. Lancet. 1989;333(8634):347–9.
Hellani A, Coskun S, Benkhalifa M, Tbakhi A, Sakati N, Al-Odaib A, et al. Multiple displacement amplification on single cell and possible PGD applications. Mol Hum Reprod. 2004;10(11):847–52.
Wells D, Sherlock JK. Strategies for preimplantation genetic diagnosis of single gene disorders by DNA amplification. Prenat Diagn. 1998;18(13):1389–401.
Natesan SA, Bladon AJ, Coskun S, Qubbaj W, Prates R, Munne S, et al. Genome-wide karyomapping accurately identifies the inheritance of single-gene defects in human preimplantation embryos in vitro. Genet Med. 2014;16(11):838–45.
Thornhill AR, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, et al. Karyomapping—a comprehensive means of simultaneous monogenic and cytogenetic PGD: comparison with standard approaches in real time for Marfan syndrome. J Assist Reprod Genet. 2015;32(3):347–56.
Natesan SA, Handyside AH, Ottolini C, Natesan SA, Taylor J, Sage K, et al. Live birth after PGD with confirmation by a comprehensive approach (karyomapping) for simultaneous detection of monogenic and chromosomal disorders. Reprod BioMed Online. 2014;29(5):600–5.
Wilton L, Thornhill A, Traeger-Synodinos J, Sermon KD, Harper JC. The causes of misdiagnosis and adverse outcomes in PGD. Hum Reprod. 2009;24(5):1221–8.
Wang, L, Cram DS, Shen J, Wang X, Zhang J, Song Z, Xu G, Li N, Fan J, Wang S, Luo Y, Wang J, Yu L, Liu J, Yao Y, Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biology of reproduction, 2014: p. biolreprod. 114.120576.
Korenberg J, Chen XN, Schipper R, Sun Z, Gonsky R, Gerwehr S, et al. Down syndrome phenotypes: the consequences of chromosomal imbalance. Proc Natl Acad Sci. 1994;91(11):4997–5001.
Treff NR, Northrop LE, Kasabwala K, Su J, Levy B, Scott RT Jr. Single nucleotide polymorphism microarray–based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil and Steril. 2011;95(5):1606–12. e2.
Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88(3):69.
Yang Z, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Mol Cytogenet. 2012;5(1):1–8.
Schoolcraft W, Surrey E, Minjarez D, Gustofson RL, Scott RT Jr, Katz-Jaffe MG. Comprehensive chromosome screening (CCS) with vitrification results in improved clinical outcome in women > 35 years: a randomized control trial. Fertil Steril. 2012;98(3):S1.
Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fert Steril. 2013;100(1):100–7. e1.
Scott RT Jr, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertil Steril. 2013;100(3):697–703.
Yan L, Huang L, Xu L, Huang J, Ma F, Zhu X, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112(52):15964–9.
Ren Y, Zhi X, Zhu X, Huang J, Lian Y, Li R, et al. Clinical applications of MARSALA for preimplantation genetic diagnosis of spinal muscular atrophy. J Genet Genomics. 2016;43(9):541–7.
Jiao Z, Zhou C, Li J, Shu Y, Liang X, Zhang M, et al. Birth of healthy children after preimplantation diagnosis of β-thalassemia by whole-genome amplification. Prenat Diagn. 2003;23(8):646–51.
Zong C, Lu S, Chapman AR, Xie XS. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338(6114):1622–6.
Zhang L, Cui X, Schmitt K, Hubert R, Navidi W, Arnheim N. Whole genome amplification from a single cell: implications for genetic analysis. Proc Natl Acad Sci. 1992;89(13):5847–51.
Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338(6114):1627–30.
Zhou X, Ren L, Meng Q, Li Y, Yu Y, Yu J. The next-generation sequencing technology and application. Protein Cell. 2010;1(6):520–36.
Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150(2):402–12.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4.
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148(5):873–85.
Xu X, Zhou YQ, Luo GX, Liao C, Zhou M, Chen PY, et al. The prevalence and spectrum of α and β thalassaemia in Guangdong Province: implications for the future health burden and population screening. J Clin Pathol. 2004;57(5):517–22.
Huang L, Ma F, Chapman A, Lu S, Xie XS. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102.
Zamani Esteki M, Dimitriadou E, Mateiu L, Melotte C, Van der Aa N, Kumar P, et al. Concurrent whole-genome haplotyping and copy-number profiling of single cells. Am J Hum Genet. 2015;96(6):894–912.
Sermon K. Novel technologies emerging for preimplantation genetic diagnosis and preimplantation genetic testing for aneuploidy, expert review of molecular diagnostics. Expert Rev Mol Diagn. 2017;17(1):71–82.
Shang W, Zhang Y, Shu M, Wang W, Li R, Chen F, et al. Comprehensive chromosomal and mitochondrial copy number profiling in human IVF embryos. Reprod BioMed Online. 2017;36:67–74. https://doi.org/10.1016/j.rbmo.2017.10.110.
Funding
The work was supported by grants from the National Natural Science Foundation of China (81370765), Guangdong Provincial Key Laboratory of Reproductive Medicine (2012A06140003), Guangzhou Science and Technology Foundation (201300000097), National Key Technologies Research and Development program (2016YFC0900100 to L.H.), Beijing Municipal Science & Technology Commission Grants (D1511000024150002 to X.S.X.), the Pilot Construction of Reproductive Clinical Research and Transformation Center of Guangzhou (155700011/201508020006), and the funding from Beijing Advanced Innovation Center for Genomics at Peking University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
X.S.X. and S.L. are cofounders of Yikon Genomics Company, Ltd.
Rights and permissions
About this article
Cite this article
Wu, H., Shen, X., Huang, L. et al. Genotyping single-sperm cells by universal MARSALA enables the acquisition of linkage information for combined pre-implantation genetic diagnosis and genome screening. J Assist Reprod Genet 35, 1071–1078 (2018). https://doi.org/10.1007/s10815-018-1158-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-018-1158-9