Abstract
Purpose
The purpose of this study was to apply next-generation sequencing (NGS) technology to identify chromosomally normal embryos for transfer in preimplantation genetic diagnosis (PGD) cycles for translocations.
Methods
A total of 21 translocation couples with a history of infertility and repeated miscarriage presented at our PGD clinic for 24-chromosome embryo testing using copy number variation sequencing (CNV-Seq).
Results
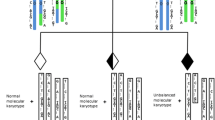
Testing of 98 embryo samples identified 68 aneuploid (69.4 %) and 30 (30.6 %) euploid embryos. Among the aneuploid embryos, the most common abnormalities were segmental translocation imbalances, followed by whole autosomal trisomies and monosomies, segmental imbalances of non-translocation chromosomes, and mosaicism. In all unbalanced embryos resulting from reciprocal translocations, CNV-Seq precisely identified both segmental imbalances, extending from the predicted breakpoints to the chromosome termini. From the 21 PGD cycles, eight patients had all abnormal embryos and 13 patients had at least one normal/balanced and euploid embryo available for transfer. In nine intrauterine transfer cycles, seven healthy babies have been born. In four of the seven children tested at 18 weeks gestation, the karyotypes matched with the original PGD results.
Conclusion
In clinical PGD translocation cycles, CNV-Seq displayed the hallmarks of a comprehensive diagnostic technology for high-resolution 24-chromosome testing of embryos, capable of identifying true euploid embryos for transfer.


Similar content being viewed by others
Abbreviations
- IVF:
-
In vitro fertilization
- NGS:
-
Next-generation sequencing
- PGD:
-
Preimplantation genetic diagnosis
- t:
-
Translocation
- WGA:
-
Whole genome amplification
References
Ogilvie CM, Braude P, Scriven PN. Successful pregnancy outcomes after preimplantation genetic diagnosis (PGD) for carriers of chromosome translocations. Hum Fertil (Camb). 2001;4:168–71.
Scriven PN, Handyside AH, Ogilvie CM. Chromosome translocations: segregation modes and strategies for preimplantation genetic diagnosis. Prenat Diagn. 1998;18:1437–49.
Munné S. Analysis of chromosome segregation during preimplantation genetic diagnosis in both male and female translocation heterozygotes. Cytogenet Genome Res. 2005;111:305–9.
Suzumori N, Sugiura-Ogasawara M. Genetic factors as a cause of miscarriage. Curr Med Chem. 2010;17:3431–7.
Munné S, Sandalinas M, Escudero T, Fung J, Gianaroli L, Cohen J. Outcome of preimplantation genetic diagnosis of translocations. Fertil Steril. 2000;73:1209–18.
Hardy K, Handyside AH. Biopsy of cleavage stage human embryos and diagnosis of single gene defects by DNA amplification. Arch Pathol Lab Med. 1992;116:388–92.
De Vos A, Van Steirteghem A. Aspects of biopsy procedures prior to preimplantation genetic diagnosis. Prenat Diagn. 2001;21:767–80.
Kokkali G, Vrettou C, Traeger-Synodinos J, Jones GM, Cram DS, Stavrou D, et al. Birth of a healthy infant following trophectoderm biopsy from blastocysts for PGD of beta-thalassaemia major. Hum Reprod. 2005;20:1855–9.
McArthur SJ, Leigh D, Marshall JT, de Boer KA, Jansen RP. Pregnancies and live births after trophectoderm biopsy and preimplantation genetic testing of human blastocysts. Fertil Steril. 2005;84:1628–36.
Fischer J, Colls P, Escudero T, Munné S. Preimplantation genetic diagnosis (PGD) improves pregnancy outcome for translocation carriers with a history of recurrent losses. Fertil Steril. 2010;94:283–9.
Fiorentino F, Spizzichino L, Bono S, Biricik A, Kokkali G, Rienzi L, et al. PGD for reciprocal and Robertsonian translocations using array comparative genomic hybridization. Hum Reprod. 2011;26:1925–35.
Treff NR, Northrop LE, Kasabwala K, Su J, Levy B, Scott Jr RT. Single nucleotide polymorphism microarray-based concurrent screening of 24-chromosome aneuploidy and unbalanced translocations in preimplantation human embryos. Fertil Steril. 2011;95:1606–12.e1-2.
Tan Y, Yin X, Zhang S, Jiang H, Tan K, Li J, et al. Clinical outcome of preimplantation genetic diagnosis and screening using next generation sequencing. Gigascience. 2014;3:30.
Munné S. Preimplantation genetic diagnosis for aneuploidy and translocations using array comparative genomic hybridization. Curr Genomics. 2012;13:463–70.
Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15:577–83.
Voet T, Vanneste E, Van der Aa N, Melotte C, Jackmaert S, Vandendael T, et al. Breakage-fusion-bridge cycles leading to inv dup del occur in human cleavage stage embryos. Hum Mutat. 2011;32:783–93.
Yin X, Tan K, Vajta G, Jiang H, Tan Y, Zhang C, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88:69.
Huang J, Yan L, Fan W, Zhao N, Zhang Y, Tang F, et al. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril. 2014;102:1685–91.
Wang L, Cram DS, Shen J, Wang X, Zhang J, Song Z, et al. Validation of copy number variation sequencing for detecting chromosome imbalances in human preimplantation embryos. Biol Reprod. 2014;91:37.
Bono S, Biricik A, Spizzichino L, Nuccitelli A, Minasi MG, Greco E, et al. Validation of a semiconductor next-generation sequencing-based protocol for preimplantation genetic diagnosis of reciprocal translocations. Prenat Diagn. 2015;35:938–44.
Fan J, Wang L, Wang H, Ma M, Wang S, Liu Z, et al. The clinical utility of next-generation sequencing for identifying chromosome disease syndromes in human embryos. Reprod Biomed Online. 2015;31:62–70.
Wang H, Wang L, Ma M, Song Z, Zhang J, Xu G, et al. A PGD pregnancy achieved by embryo copy number variation sequencing with confirmation by non-invasive prenatal diagnosis. J Genet Genomics. 2014;41:453–6.
Ruttanajit T, Chanchamroen S, Cram DS, Sawakwongpra K, Suksalak W, Leng X, et al. Detection and quantitation of chromosomal mosaicism in human blastocysts using copy number variation sequencing. Prenat Diagn. 2015;36:154–62.
Liang D, Peng Y, Lv W, Deng L, Zhang Y, Li H, et al. Copy number variation sequencing for comprehensive diagnosis of chromosome disease syndromes. J Mol Diagn. 2014;16:519–26.
Wang Y, Chen Y, Tian F, Zhang J, Song Z, Wu Y, et al. Maternal mosaicism is a significant contributor to discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Clin Chem. 2014;60:251–9.
Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenat Diagn. 2013;33:409–15.
Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95.
Tibshirani R, Wang P. Spatial smoothing and hot spot detection for CGH data using fused lasso. Biostatistics. 2008;9:18–29.
Handyside AH. 24-chromosome copy number analysis: a comparison of available technologies. Fertil Steril. 2013;100:595–602.
Li N, Wang L, Wang H, Ma M, Wang X, Li Y, et al. The performance of whole genome amplification methods and next-generation sequencing for pre-implantation genetic diagnosis of chromosomal abnormalities. J Genet Genomics. 2015;42:151–9.
Fiorentino F, Biricik A, Bono S, Spizzichino L, Cotroneo E, Cottone G, et al. Development and validation of a next-generation sequencing-based protocol for 24-chromosome aneuploidy screening of embryos. Fertil Steril. 2014;101:1375–82.
Wells D, Kaur K, Grifo J, Glassner M, Taylor JC, Fragouli E, et al. Clinical utilisation of a rapid low-pass whole genome sequencing technique for the diagnosis of aneuploidy in human embryos prior to implantation. J Med Genet. 2014;51:553–62.
Acknowledgments
The study was supported by a grant awarded to Yuanqing Yao by the Key Program of the “Twelfth Five-Year Plan” of the People’s Liberation Army (BWS11J058) and the National High Technology Research and Development Program (SS2015AA020402). Li Wang was supported by a grant from the China Postdoctoral Science Foundation.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical approval
Clinical PGD was conducted at the Department of Obstetrics and Gynecology, Chinese PLA General Hospital, Beijing, with approval from the Ethics Committee of the Chinese PLA General Hospital (S2013-092-02). All patients provided written informed consent for IVF and PGD.
Additional information
Capsule
Next-generation sequencing offers increased chromosome resolution for comprehensive preimplantation genetic diagnosis of translocations.
Wenke Zhang, Ying Liu and Li Wang contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 37 kb)
Rights and permissions
About this article
Cite this article
Zhang, W., Liu, Y., Wang, L. et al. Clinical application of next-generation sequencing in preimplantation genetic diagnosis cycles for Robertsonian and reciprocal translocations. J Assist Reprod Genet 33, 899–906 (2016). https://doi.org/10.1007/s10815-016-0724-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-016-0724-2