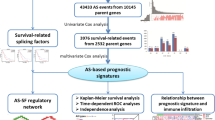
Abstract
Purpose
Alternative splicing (AS) events were reportedly associated with the development of multiple cancers. The study was designed to provide a comprehensive analysis of AS events and explore their potential prognostic value in uveal melanoma (UM).
Methods
The prognostic AS events, identified based on the data of 80 UM patients obtained from The Cancer Genome Atlas, were further screened and analyzed for construction of prognostic signatures by using LASSO regression and multivariate Cox model. Kaplan–Meier survival analysis was used to evaluate the prognostic value. The AS events-related functional pathways were explored by gene set enrichment analysis (GSEA). The difference between two subgroups in terms of treatment options was investigated. The regulatory network between prognostic AS events and splicing factors (SFs) was then constructed.
Results
A total of 1014 AS events were identified as prognostic AS events. Five prognostic AS events were involved in the construction of prognostic signatures, including AKAP2/87175/AP, RGMA/32575/ES, DNASE1L1/90581/ES, BIN1/55198/ES and ERCC2/50430/AT. UM patients were then divided into two subgroups. Prognostic AS signatures had an excellent performance in predicting the survival of UM patients, with an area under curve (AUC) of 0.962. GSEA results suggested several splicing-associated mechanisms, including cellular metabolic process and apoptosis. Low-risk subgroup could be more sensitive to drugs. A higher expression of immune checkpoint genes was observed in high-risk group than in low-risk group. SFs-AS regulatory network also revealed significant association between AS events and SFs.
Conclusions
Aberrant AS events in UM patients might serve as prognostic predictors.








Similar content being viewed by others
Abbreviations
- AA:
-
Alternate acceptor
- AD:
-
Alternate donor
- AP:
-
Alternate promoter
- AT:
-
Alternate terminator
- AS:
-
Alternative splicing
- AUC:
-
Area under curve
- ES:
-
Exon skip
- GSEA:
-
Gene set enrichment analysis
- HR:
-
Hazard ration
- LASSO:
-
The least absolute shrinkage and selection operator
- ME:
-
Mutually exclusive exon
- PSI:
-
Percent Spliced In
- UM:
-
Uveal melanoma
- RI:
-
Retained intron
- RNA-seq:
-
RNA sequencing
- ROC:
-
Receiver operating characteristic
- SFs:
-
Splicing factors
- TCGA:
-
The Cancer Genome Atlas
References
Singh AD, Turell ME, Topham AK (2011) Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology 118(9):1881–1885. https://doi.org/10.1016/j.ophtha.2011.01.040
Kivela T (2009) The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br J Ophthalmol 93(9):1129–1131. https://doi.org/10.1136/bjo.2008.150292
Kaliki S, Shields CL (2017) Uveal melanoma: relatively rare but deadly cancer. Eye (Lond) 31(2):241–257. https://doi.org/10.1038/eye.2016.275
Shields CL, Kaliki S, Furuta M, Fulco E, Alarcon C, Shields JA (2013) American Joint Committee on Cancer classification of posterior uveal melanoma (tumor size category) predicts prognosis in 7731 patients. Ophthalmology 120(10):2066–2071. https://doi.org/10.1016/j.ophtha.2013.03.012
Shields CL, Furuta M, Thangappan A, Nagori S, Mashayekhi A, Lally DR, Kelly CC, Rudich DS, Nagori AV, Wakade OA (2009) Metastasis of uveal melanoma millimeter-by-millimeter in 8033 consecutive eyes. Arch Ophthalmol 127(8):989–998. https://doi.org/10.1001/archophthalmol.2009.208
Khoja L, Atenafu EG, Suciu S, Leyvraz S, Sato T, Marshall E, Keilholz U, Zimmer L, Patel SP, Piperno-Neumann S, Piulats J, Kivela TT, Pfoehler C, Bhatia S, Huppert P, Van Iersel LBJ, De Vries IJM, Penel N, Vogl T, Cheng T, Fiorentini G, Mouriaux F, Tarhini A, Patel PM, Carvajal R, Joshua AM (2019) Meta-analysis in metastatic uveal melanoma to determine progression free and overall survival benchmarks: an international rare cancers initiative (IRCI) ocular melanoma study. Ann Oncol 30(8):1370–1380. https://doi.org/10.1093/annonc/mdz176
Smit KN, Jager MJ, de Klein A, Kiliҫ E (2019) Uveal melanoma: towards a molecular understanding. Prog Retin Eye Res. https://doi.org/10.1016/j.preteyeres.2019.100800
Damato B, Eleuteri A, Taktak AF, Coupland SE (2011) Estimating prognosis for survival after treatment of choroidal melanoma. Prog Retin Eye Res 30(5):285–295. https://doi.org/10.1016/j.preteyeres.2011.05.003
van den Bosch T, van Beek JG, Vaarwater J, Verdijk RM, Naus NC, Paridaens D, de Klein A, Kilic E (2012) Higher percentage of FISH-determined monosomy 3 and 8q amplification in uveal melanoma cells relate to poor patient prognosis. Invest Ophthalmol Vis Sci 53(6):2668–2674. https://doi.org/10.1167/iovs.11-8697
Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S (2013) SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 3(10):1122–1129. https://doi.org/10.1158/2159-8290.CD-13-0330
Chen M, Manley JL (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol 10(11):741–754. https://doi.org/10.1038/nrm2777
Hegyi H, Kalmar L, Horvath T, Tompa P (2011) Verification of alternative splicing variants based on domain integrity, truncation length and intrinsic protein disorder. Nucleic Acids Res 39(4):1208–1219. https://doi.org/10.1093/nar/gkq843
Park E, Pan Z, Zhang Z, Lin L, Xing Y (2018) The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet 102(1):11–26. https://doi.org/10.1016/j.ajhg.2017.11.002
Lee SC, Abdel-Wahab O (2016) Therapeutic targeting of splicing in cancer. Nat Med 22(9):976–986. https://doi.org/10.1038/nm.4165
Tomczak K, Czerwinska P, Wiznerowicz M (2015) The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19(1A):A68-77. https://doi.org/10.5114/wo.2014.47136
Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J (2016) TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 44(D1):D1018-1022. https://doi.org/10.1093/nar/gkv1288
Schafer S, Miao K, Benson CC, Heinig M, Cook SA, Hubner N (2015) Alternative splicing signatures in RNA-seq Data: Percent Spliced in (PSI). Curr Protoc Hum Genet 87:111611–111614. https://doi.org/10.1002/0471142905.hg1116s87
Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, Zhou Q, Xing Y (2014) rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc Natl Acad Sci USA 111(51):E5593-5601. https://doi.org/10.1073/pnas.1419161111
Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H (2014) UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph 20(12):1983–1992. https://doi.org/10.1109/TVCG.2014.2346248
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J Roy Stat Soc: Ser B Methodol 58(1):267–288. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102(43):15545–15550. https://doi.org/10.1073/pnas.0506580102
Tilford CA, Siemers NO (2009) Gene set enrichment analysis. In: Protein networks and pathway analysis. Springer, Berlin, pp 99–121. doi:https://doi.org/10.1007/978-1-60761-175-2_6
Piva F, Giulietti M, Nocchi L, Principato G (2009) SpliceAid: a database of experimental RNA target motifs bound by splicing proteins in humans. Bioinformatics 25(9):1211–1213. https://doi.org/10.1093/bioinformatics/btp124
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Shields CL, Manalac J, Das C, Ferguson K, Shields JA (2014) Choroidal melanoma: clinical features, classification, and top 10 pseudomelanomas. Curr Opin Ophthalmol 25(3):177–185. https://doi.org/10.1097/ICU.0000000000000041
Seftor EA, Meltzer PS, Kirschmann DA, Pe’er J, Maniotis AJ, Trent JM, Folberg R, Hendrix MJ (2002) Molecular determinants of human uveal melanoma invasion and metastasis. Clin Exp Metastasis 19(3):233–246. https://doi.org/10.1023/a:1015591624171
Harbour JW (2012) The genetics of uveal melanoma: an emerging framework for targeted therapy. Pigment Cell Melanoma Res 25(2):171–181. https://doi.org/10.1111/j.1755-148X.2012.00979.x
Augsburger JJ, Gamel JW (1990) Clinical prognostic factors in patients with posterior uveal malignant melanoma. Cancer 66(7):1596–1600. https://doi.org/10.1002/1097-0142(19901001)66:7%3c1596::aid-cncr2820660726%3e3.0.co;2-6
De Cruz Jr POL, Specht CS, McLean IW (1990) Lymphocytic infiltration in uveal malignant melanoma. Cancer 65(1):112–115. https://doi.org/10.1002/1097-0142(19900101)65:1%3c112::aid-cncr2820650123%3e3.0.co;2-x
Folberg R, Pe’er J, Gruman LM, Woolson RF, Jeng G, Montague PR, Moninger TO, Yi H, Moore KC (1992) The morphologic characteristics of tumor blood vessels as a marker of tumor progression in primary human uveal melanoma: a matched case-control study. Hum Pathol 23(11):1298–1305. https://doi.org/10.1016/0046-8177(92)90299-i
Bornfeld N, Prescher G, Becher R, Hirche H, Jöckel K, Horsthemke B (1996) Prognostic implications of monosomy 3 in uveal melanoma. The Lancet 347(9010):1222–1225. https://doi.org/10.1016/s0140-6736(96)90736-9
White VA, Chambers JD, Courtright PD, Chang WY, Horsman DE (1998) Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer Interdiscipl Int J Am Cancer Soc 83(2):354–359
Horsman DE, White VA (1993) Cytogenetic analysis of uveal melanoma consistent occurrence of monosomy 3 and trisomy 8q. Cancer 71(3):811–819. https://doi.org/10.1002/1097-0142(19930201)71:3%3c811::aid-cncr2820710325%3e3.0.co;2-f
Sisley K, Rennie IG, Parsons MA, Jacques R, Hammond DW, Bell SM, Potter AM, Rees RC (1997) Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosom Cancer 19(1):22–28. https://doi.org/10.1002/(sici)1098-2264(199705)19:1%3c22::aid-gcc4%3e3.0.co;2-2
Onken MD, Worley LA, Tuscan MD, Harbour JW (2010) An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn 12(4):461–468. https://doi.org/10.2353/jmoldx.2010.090220
Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, Aaberg TM Jr, Altaweel MM, Bardenstein DS, Finger PT (2012) Collaborative ocular oncology group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 119(8):1596–1603. https://doi.org/10.1016/j.ophtha.2012.02.017
Oltean S, Bates DO (2014) Hallmarks of alternative splicing in cancer. Oncogene 33(46):5311–5318. https://doi.org/10.1038/onc.2013.533
Venables JP (2004) Aberrant and alternative splicing in cancer. Cancer Res 64(21):7647–7654. https://doi.org/10.1158/0008-5472.CAN-04-1910
Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI (2016) Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 35(19):2413–2427. https://doi.org/10.1038/onc.2015.318
Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S, Stern MH, Marais R (2013) SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 3(10):1122–1129. https://doi.org/10.1158/2159-8290.CD-13-0330
Yavuzyigitoglu S, Koopmans AE, Verdijk RM, Vaarwater J, Eussen B, van Bodegom A, Paridaens D, Kilic E, de Klein A, Rotterdam Ocular Melanoma Study G (2016) Uveal melanomas with SF3B1 mutations: a distinct subclass associated with late-onset metastases. Ophthalmology 123(5):1118–1128. https://doi.org/10.1016/j.ophtha.2016.01.023
Luengo A, Gui DY, Vander Heiden MG (2017) Targeting metabolism for cancer therapy. Cell Chem Biol 24(9):1161–1180. https://doi.org/10.1016/j.chembiol.2017.08.028
Pavlova NN, Thompson CB (2016) The emerging hallmarks of cancer metabolism. Cell Metab 23(1):27–47. https://doi.org/10.1016/j.cmet.2015.12.006
Zhang Y, Yang Y, Chen L, Zhang J (2014) Expression analysis of genes and pathways associated with liver metastases of the uveal melanoma. BMC Med Genet 15(1):29. https://doi.org/10.1186/1471-2350-15-29
Goh AY, Layton CJ (2016) Evolving systemic targeted therapy strategies in uveal melanoma and implications for ophthalmic management: a review. Clin Exp Ophthalmol. https://doi.org/10.1111/ceo.12688
Dong H, Jo J, Hyoung K, Jeong H (2019) Targeting tyrosine kinases for treatment of ocular tumors. Arch Pharmacal Res. https://doi.org/10.1007/s12272-018-1094-3
Ericsson C, Seregard S, Bartolazzi A, Levitskaya E, Ferrone S, Kiessling R, Larsson O (2001) Association of HLA class I and class II antigen expression and mortality in uveal melanoma. Invest Ophthalmol Vis Sci 42(10):2153–2156. https://doi.org/10.1097/00004397-200110000-00022
Souri Z, Wierenga A, Mulder A, Jochemsen AG, Jager MJ (2019) HLA expression in uveal melanoma: an indicator of malignancy and a modifiable immunological target. Cancers 11(8):1132. https://doi.org/10.3390/cancers11081132
Grosso AR, Martins S, Carmo-Fonseca M (2008) The emerging role of splicing factors in cancer. EMBO Rep 9(11):1087–1093. https://doi.org/10.1038/embor.2008.189
Zhang J, Manley JL (2013) Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov 3(11):1228–1237. https://doi.org/10.1158/2159-8290.CD-13-0253
Acknowledgements
The authors would like to thank the TCGA research network (http://cancergenome.nih.gov) for their efforts in making this data publicly available.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xie, X., Zheng, X., Xie, T. et al. Identification of prognostic alternative splicing signatures in uveal melanoma. Int Ophthalmol 41, 1347–1362 (2021). https://doi.org/10.1007/s10792-021-01699-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10792-021-01699-z