Abstract
We explore the feasibility of automatically identifying sentences in different MEDLINE abstracts that are related in meaning. We compared traditional vector space models with machine learning methods for detecting relatedness, and found that machine learning was superior. The Huber method, a variant of Support Vector Machines which minimizes the modified Huber loss function, achieves 73% precision when the score cutoff is set high enough to identify about one related sentence per abstract on average. We illustrate how an abstract viewed in PubMed might be modified to present the related sentences found in other abstracts by this automatic procedure.
Similar content being viewed by others
1 Introduction
Search engines respond to a user query by producing a list of documents from a given collection, ordering the list according to the user’s supposed information need. However, even the most relevant documents will contain some portions of greater interest to the user, and other portions of little or no interest. This may explain why for example, when querying over full text collections, retrieval performance can be improved by segmenting documents into sections or paragraphs, and matching or retrieving passages rather than full documents (Hearst and Plaunt 1993; Lin 2009), although mixed results have been reported for matching queries at the sentence level (Ko et al. 2002; Lu et al. 2009; Salton and Buckley 1991).
PubMedFootnote 1 is the search engine for articles in MEDLINE maintained at the National Library of Medicine (Sayers et al. 2009; Wilbur 2005). When a user selects an article to view, a list of related articles may also appear alongside its other details. Related articles are pre-computed using a topic-based model that measures the size of overlapping subject matter of two articles (Lin and Wilbur 2007). Although the related articles feature is popular (20% of user sessions involve viewing a related article) it assumes that users are primarily interested in articles that have maximal overlapping subject matter. The goal of this paper is to explore an alternative method of finding related content, to address the needs of users interested in a particular sentence in an article by finding other related sentences. We assume users would be interested in other occurrences of the same sentence, a restatement of the sentence, or any sentence that makes a closely related assertion.
We used vector space models to estimate the relatedness of sentences. In addition to the tf-idf formula suggested by (Salton and Buckley 1991), we also adapted several well known retrieval functions, such as the Dice coefficient, cosine similarity, and bm25. But fixed formulas give only one possibility for term weights in a vector space model, and theoretically it should be possible to use machine learning to find optimal term weights.
Machine learning has been applied in an analogous setting of information retrieval. The goal of learning to rank is to use machine learning to obtain retrieval scoring functions for optimal ranking of query results (Joachims et al. 2007). That research has been limited in the past by the availability of test data, and much of the effort has been focused on effective learning algorithms to meet the unique challenges. The focus may shift now that the LETOR corpus has emerged as a community benchmark dataset (Liu et al. 2007), and with methods for automated annotation derived from user clickthrough data (Joachims 2002).
As with learning to rank, the biggest challenge to machine learning of related sentences is the availability of a usable corpus. To our knowledge, no datasets of related sentences have been discussed in the research literature. We claim that a productive corpus must be large enough to contain many examples of related sentences on a variety of different topics, and that manual annotation of such a large corpus of sentence pairs is not feasible. Fortunately, there is an ideal solution to this problem. Firstly, the MEDLINE database contains a large number of sentences (available in article abstracts) in many different subject areas. And secondly, sentences are likely to be related if they are adjacent sentences from the same MEDLINE abstract, and unrelated if they are from different randomly selected abstracts. Thus it is possible to automatically assemble a very large training corpus of sentence pairs from MEDLINE.
Our ability to detect the relatedness of two sentences is dependent on their sharing words or parts of words. We do not use a thesaurus or dictionary. We have found words and portions of words to be the most useful features in our approach. Such features are used in our application of the standard information retrieval formulas that we test, as well as in our machine learning. In a minor departure from the vector space model, we also learned weights for terms that appear in only one sentence, which tend to be negative and reflect the evidence for two sentences to be unrelated when one sentence contains the word and the other does not.
Having a large annotated corpus of related sentences makes it possible to apply machine learning to obtain optimal weights for a vector space model, and to compare the results with traditional vector space models. Indeed, we found that the learned model was significantly better than any of the traditional models at recognizing related sentences. This result means that we can distinguish pairs of sentences that were adjacent in an abstract from pairs of sentences randomly selected from different abstracts at a high level. However, this is not our ultimate goal. Our goal is to apply the learning for a given query sentence to find related sentences in other documents. We performed a modest manual evaluation which shows that the model from machine learning also gives significantly better results at this task. In summary, our contribution is a first use of a large automatically generated training corpus for related sentences, and the demonstration that the model learned from this corpus outperforms traditional vector space methods at finding related sentence pairs in different documents.
In the remainder of the paper, Sect. 2 describes our corpus of related sentence pairs, the baseline and machine learning methods used to detect relatedness, and our approaches to evaluating the results. Section 3 gives the results of the baseline and machine learning methods, followed by a manual comparison of the best of each, and further evaluation of the best method. A potential application is illustrated in Sect. 4. Limitations of the concept of relatedness are discussed in Sect. 5. We conclude with a summary in Sect. 6.
2 Methods
We will now describe the corpus that we created for this study, the various methods of relatedness detection that we considered, and the techniques we used to evaluate them.
2.1 Corpus
We began with a snapshot of MEDLINE, taken in June, 2008, from which we extracted the abstracts, and divided them into sentences using the MedPost part of speech tagger (Smith et al. 2004). There were a total of 73,812,862 sentences at this stage.
We wanted to ignore sentences which contained only generic words. To do this, we used the result of a previous study (Kim and Wilbur 2001) which for each word w defined a strength of context score, s 1 (w), that serves as a measure of how strongly w is related to its context. The authors of that paper provided us with a set of s 1-scores for 535,533 words, from which we selected 153,737 high scoring content words by arbitrarily requiring s 1(w) > 160.0. Sentences were eliminated from consideration if they did not contain at least one of these content words, resulting in a total of 37,371,346 sentences.
Among these remaining sentences, if two were found to be in the same abstract and adjacent to each other, then they were added to the corpus of sentence pairs. In keeping with our relatedness hypothesis, these were all annotated as positive examples of related sentence pairs. This resulted in 28,771,427 related sentence pairs from 6,552,370 different abstracts, or about 5.7 sentences per content-containing abstract.
To obtain negative examples, we took this same list of related sentence pairs and randomly permuted the second sentence of all the pairs. The resulting pairs, none of which were from the same abstract, were added to the corpus as negative examples of related sentence pairs. All together, our corpus consisted of 57,542,854 sentence pairs, half of which were positive.
One-third of the corpus was designated as the test set, resulting in 19,180,952 sentence pairs. The remaining 38,361,902 sentence pairs were designated as the training set. In both the training and test sets, half of the sentence pairs were from the positive class and half from the negative class. In addition, to study feature selection and parameter tuning, we selected a tuning set of about 5 million sentence pairs such that half were annotated positive, and half negative. The tuning set was chosen to consist of all abstracts produced by a “cardiovascular disease” PubMed query to insure that it would contain a large number of related sentences in the same topic area.
2.2 Features
Each pair of sentences in the corpus was represented in the database by three sets of features: one set for each sentence, and a set for the pair as a whole, derived in a simple way from the two sets of sentence features. Our baseline methods used the features associated with sentences, while our machine learning methods used the features associated with pairs. We now describe the features in detail.
Each sentence was divided into tokens by breaking on all non-alphanumeric characters, retaining only strings that contained at least one letter, and excluding a set of 313 common stop words (Wilbur and Sirotkin 1992). These strings were mapped to lower case and designated sentence word features, or type W features. Substrings of the type W features with varying lengths (which also contained at least one letter) were designated sentence substring features, or type S features. The number of times that each sentence feature occurred in a sentence was also recorded (some of the baseline methods used this data).
Features associated with a pair of sentences were defined by comparing the two sets of sentence features. Those sentence features that appeared in both sets were designated as pair intersection features, or type I features. Those that occurred in only one of the sets were designated as pair disjoint features, or type D features. Thus, each feature of a sentence pair was described by a string (which appears in at least one of the sentences), a sentence feature type (W or S) and a pair feature type (D or I).
Based on a preliminary study on the tuning set, the best performance was achieved when type S features of length 2–6 were combined with type W features. With this definition, the 37,371,346 sentences in the corpus had an average of 16.1 type W features and 442.7 type S features per sentence. The 57,542,854 sentence pairs in the corpus had an average of 80.7 type I features and 630.1 type D features per pair.
2.3 Baseline methods
We tested whether machine learning was more effective at relatedness detection than traditional methods of measuring content overlap. Several relatedness measures commonly used in information retrieval were identified and used as baseline methods. Each of these methods is defined by a formula that takes as its input two sets of sentence features, and produces a number that measures the amount of overlap in the two sets. The formulas for all baseline methods are given in Table 1.
The Jaccard index and Dice coefficient are the simplest methods and have a long history in information retrieval (van Rijsbergen 1979). Their formulas depend on the number of distinct features in each set, the size of the intersection and the size of the union. However, the two measures always produce the same ranking, as can be verified by elementary algebra, and so we only report the results for the Dice coefficient (D).
Three of the baseline methods were based on the OKAPI bm25 formula (Robertson and Spark Jones 1994), originally designed to measure the degree of relevance of a document given a keyword query. The bm25 similarity score is the sum of term frequency tf t,S times an inverse document frequency df t for all terms t in a query (formulas shown in Table 1). But applying this formula directly to our situation would require us to designate one sentence as a query sentence and the other as a document. To preserve symmetry, we sum over all terms common to both sentences, and instead of the term frequency we use the product of term frequencies (O 1), geometric mean of term frequencies (O 2), or 1 (O 3).
In Automatic Text Structuring (Salton and Buckley 1991), sentences were compared for similarity using a dot product of weight vectors called atn. The weights were defined for terms in a sentence as a product of a term frequency formula and inverse document frequency formula. We used this same formula for a baseline method denoted A.
Two baseline methods were based on the TextTiling algorithm (Hearst 1993), originally developed to partition a document into coherent sections. That paper defines tf.idf for a term to be the frequency of a term within a document (tf) divided by the frequency of a term throughout the whole collection (idf; both formulas shown in Table 1). Each document, in our case a sentence, is associated with a vector of tf.idf defined this way, and the TextTiling method (T 1) uses the cosine similarity of these vectors. The dot product of tf.idf was also used as a method (T 2).
Finally, generalizing the tf.idf dot product formula by taking tf = 1, we define simple measures of similarity equal to the sum of the inverse document frequency raised to a power e (I e for e = 0.5, 1, 1.5, 2, 3).
2.4 Machine learning algorithm
The corpus of sentence pairs, with pair features derived from the words and strings from each sentence, and annotated for relatedness, makes it possible to apply machine learning to identify related sentences. Machine learning determines a numeric value for each feature, called a weight. For any given pair of sentences, the sum of weights over the associated pair features is used to predict whether the pair is related.
We trained on the training set with Bayesian and Huber machine learning. Definitions are given in Table 2. In all cases, we used only those features that appeared in two or more sentence pairs in the training set.
2.4.1 Baysian machine learning
Naïve Bayes machine learning determines weights from the training set on the assumption that the features are independent given the class prediction (Langley 1996; Wilbur 2000). The Bayesian weight of a feature t is defined as
where p t is the proportion of positive instances that contain the feature t, and q t is the proportion of negative instances that contain t. Note that the Bayesian weight for a given feature is independent of all other features.
2.4.2 Huber machine learning
We also performed Huber machine learning, which is a variant of support vector machines (SVM; Joachims 2006; Vapnik 1998). This method determines feature weights that minimize the modified Huber cost, an explicit function that substitutes a modified Huber loss function in place of the hinge loss function traditionally used in SVM learning (Zhang 2004; Zou et al. 2008).
To define Huber cost, let T denote the size of the training set, let the binary feature vector of the ith pair in the training set be denoted by X i , and let y i = 1 if the pair is annotated as positive and y i = −1 otherwise. Let w denote a vector of feature weights, of the same length as X i , let θ denote a threshold parameter, and let \( \lambda \) denote a regularization parameter. Then the cost function is given by:
where the function h is the modified Huber loss function:
The values of the parameters (w and θ) which minimize C can be determined using a gradient descent algorithm. The regularization parameter \( \lambda \) is computed from the training set with the formula:
where \( \left\langle {\left| x \right|} \right\rangle \) is the average Euclidean norm of the feature vectors in the training set, and \( \lambda^{\prime} \) must be chosen.Footnote 2 We optimized \( \lambda^{\prime} \) on the tuning set by examining powers of 10, and arrived at \( \lambda^{\prime} = 10^{ - 7} \), which was used to determine \( \lambda \) for all Huber training on the larger corpus.
2.5 Evaluation
We evaluated the relatedness scoring functions in two complementary ways, break-even precision and manual evaluation.
2.5.1 Precision-recall break-even
For each relatedness scoring function, the sentence pairs in the test set were scored and precision-recall break-even (BE) was computed (see Table 3). The BE is the precision at the point in the precision-recall curve where precision equals recall. This is equivalent to the precision when the number retrieved is equal to the number of positive instances in the test set. And since the numbers of positive and negative instances were equal in our corpus, it is equivalent to the precision at the median score.
2.5.2 Manual evaluation
Break-even measures the proportion of related sentences in the top half of scores in the test set. But the practical goal of our research is to find sentences that would be humanly judged to be related to a given sentence, and our test set does not specifically measure this. We therefore needed to evaluate the relatedness of the top scoring match for randomly selected query sentences. If M is a scoring method, we define the M-match of a query sentence X to be
where the maximization is performed over all sentences y in MEDLINE that are not in the same abstract as X.
The authors independently rated the similarity of 1,000 randomly selected query sentences to their corresponding M-match, for several methods M. Each similarity rating was a subjective estimate of the probability that a reader would consider the two sentences to be similar, using a scale from 0 to 4. The interpretation of the assigned rating was 0 for definitely unrelated, 1 for probably unrelated, 2 for possibly related, 3 for probably related, and 4 for definitely related. In performing evaluations, the order of presentation was randomized and all information about the source of the sentences (including the method and score) was suppressed.
Judgments of the relatedness of documents generally do not require an expert on those documents. In a highly technical area, one might desire some level of expertise. But previous research shows that some untrained workers give more accurate relevance judgments than subject experts, and that pooling the judgments of untrained workers actually outperforms the judgments of individual subject experts (Wilbur 1998). In our case, both authors have years of experience with the biological literature, and the second author has an MD degree.
2.5.3 McNemar statistic
We used the McNemar statistic to test significance of the difference between two classification methods (Dietterich 1998). For two numeric measures, B and C (which may be binary) given on a population, the statistic is defined as:
where b is the number of times B > C, and c is the number of times where C > B. This statistic is approximately \( \chi^{2} \) with 1 degree of freedom.
2.6 Computing resources
Our computer hardware consisted of 64 bit Intel Xeon dual and quad core hosts running Linux version 2.6.5. For multi-processing, we used a shared grid of 175 hosts, with a total of 1,164 cores and 4G of RAM each, managed with the Sun Grid Engine (SGE 6.2u1).
The baseline scoring of the entire corpus of sentence pairs was completed in 1,000 parallel processes. For each pair, all of the baseline methods were calculated at once, at an average time of 35.1 pairs per second. Naïve Bayes training and testing took about 6 h on a single processor.
For Huber machine learning, the cost function was minimized using an iterative gradient descent algorithm. Because there was a very large number of training instances, and each instance had around 700 features, it was not feasible to perform the minimization unless all of the required training instances remained in resident memory. And because the total size of this data was over 160 gigabytes, it was not feasible to run it on a single computer. Therefore, in order to train more quickly and to get around the resident memory limit, we developed a multiprocessing version of the algorithm. The minimization was completed in 5,440 iterations, with the gradient computed in parallel on 64 cores, averaging 73.7 s per iteration for about 4.7 days of elapsed time.
To find the M-match for each query sentence, all MEDLINE sentences were scored (using the scoring method M) and sorted using a single processor. The elapsed time for scoring each sentence was approximately 10.7 min regardless of the scoring method.
3 Results
Here we summarize the results with break-even score (BE) and manual evaluation.
3.1 Break-even
For each of the baseline and machine learning methods, the sentence pairs in the test set were scored and the BE calculated. These results are shown in Table 3. The BE was greater than 84% for all methods, confirming the self-evident hypothesis that two sentences in the same abstract are much more likely to use the same or lexically related words than a random pair of sentences.
Among the baseline methods, the Dice coefficient (D) had the lowest BE (84.12), which may reflect that this method treats all features with equal weight. With the OKAPI bm25 method, multiplying the tf (O 1) gave a BE of 88.34, but lower BE was observed with the geometric mean (O 2, 87.73) and with tf = 1 (O 3, 86.89). With the TextTiling method, the standard cosine similarity (T 1) gave a BE of 85.35, while the simpler tf.idf dot product (T 2) gave 88.69. The idf methods gave a modal response from 87.23 for I 0.5 to a maximum of 88.77 for I 1.5 and back down to 88.39 for I 3. The BE of the idf I 1.5 method was very close to the more complicated OKAPI bm25 (O 1), and was nevertheless the highest BE of all baseline methods. The I 1.5 method was used for subsequent comparison to the machine learning methods.
The machine learning methods did well relative to the baseline methods. Naïve Bayes (B) had a BE of 88.69, which is better than all but two of the baseline methods (one apparent tie differs in the next digit). Huber machine learning (H) had the highest BE of all methods at 91.08.
Because the number of instances in the test set was very large (over 19 million), very small differences in precision can be expected to be statistically significant. Indeed, the p-values for the differences in BE were negligible for all comparisons (by the McNemar test).
3.2 Manual comparison of I 1.5, B, and H
All of the differences in BE were statistically significant, yet they do not predict how the methods perform when looking at their top scoring matches for a given sentence. To determine this, we compared the top scoring sentences for the best baseline method (I 1.5) and both machine learning methods (B and H). For each pair of methods, we randomly selected 200 query sentences which had a different match for the two methods. The resulting 1,200 sentence pairs were judged by both evaluators, as described in Sect. 2.5.2. The two judgments for each sentence pair were averaged as the human standard. The average rating for all I 1.5-matches was 1.66, for B-matches 1.75, and for H-matches 1.92. A pairwise comparison of these three methods is summarized in Table 4. A significant number of query sentences had H-matches that were judged more relevant than either of the other methods (p = 0.0087 for B-matches and p = 0.021 for I 1.5-matches). Also, a greater number of query sentences had B-matches that were judged more relevant than the corresponding I 1.5-match (p = 0.091, not statistically significant).
Because there is some subjectivity in deciding whether sentences are related, we did not expect the inter-annotator agreement to be high (Artstein and Poesio 2008). And since the judgments were not used in training, we do not believe this was an issue. Yet it may be of interest that there was moderate inter-annotator agreement on whether one of the matches was more relevant than the other (Cohen’s κ = 0.4537, computed over 600 comparisons).
3.3 Manual evaluation of H
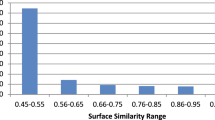
We manually rated the H-matches of 1,000 randomly selected query sentences, also following the procedure described in Sect. 2.5.2. The distribution of the average ratings is shown in Fig. 1. There was a near uniform distribution of judgments, with 481 out of 1,000 pairs (48.1%) receiving an average rating >2.
Next, we looked at the H-score to select pairs that are more likely to be related. The distribution of H-scores for the 1,000 pairs is shown in Fig. 2, grouped by the integer part of the score. The average H-score was 5.01, with quartiles 3.58, 4.55, and 5.92. The comparison of H-score and average rating is shown in Fig. 3, grouped the same as Fig. 2. The graph shows that as the H-score increases, the proportion of H-matches with high rating also increases. The number of pairs that received an average rating >2 was 409/750 (54.5%) for H ≥ 3.58, 303/500 (60.6%) for H ≥ 4.55, and 174/250 (69.6%) for H ≥ 5.92.
Plot of proportion of average rating by H-score (grouping same as Fig. 2) for 1,000 random query sentences. The vertical axis is the proportion of H-pairs in the group having the given average rating
The inter-annotator agreement (Artstein and Poesio 2008) on the judgment of the 1,000 sentence pairs was fair (κ = 0.2666), based on a five category comparison (0–4). The correlation of ratings of the two evaluators on the set of 1,000 sentences was 0.6067 (p < 10−6, determined by two-sided bootstrap). There was moderate agreement for whether a sentence pair received a rating >2 (κ = 0.4538).
4 Application
An algorithm that identifies related sentences could be used in a query setting to recommend further reading. We illustrate how this might be implemented in PubMed. Let us call an H-match related if the average human rating is >2 (this requires that at least one of the judgments was a 3 or 4). Since abstracts have an average number of sentences between 5 and 6, we chose a score cutoff H ≥ 6.4 which selects 20% of sentences, or about one per abstract, on average. At this level, the manual judgment found 148 out of 200 pairs (73%) with average rating >2. Since there were a total of 481 matches with this average rating, we expect to find approximately 148/481 (31%) of all related matches in this way.
We exhibit the related sentences for a recent MEDLINE article on an aspect of molecular biology in Alzheimer’s disease (Ma et al. 2009). Figure 4 shows a modified PubMed view of the abstract together with matching sentences. Of the 12 sentences in the abstract, 3 of them had matches with H ≥ 6.4; these are shown in boxes along with additional information about the match. All of the matches are clearly in the same subject area as the original (amyloid deposition in Alzheimer’s disease), and one of them is from an article that is actually cited in the bibliography of the paper. This also illustrates how related sentences define explicit and specific relationships to other articles, giving the user more information to guide further exploration. The matches shown are very specific and technical, and indicate different directions that a user could choose to explore.
Abstract from MEDLINE (Ma et al. 2009) with sentences highlighted that were found to have matching sentences elsewhere in MEDLINE with H ≥ 6.4. The three H-matches are shown in the shaded boxes below their corresponding query sentences
5 Limitations
The very concept of relatedness in meaning at the sentence level is theoretically imprecise due to loss of context and variability in the reader’s focus of attention and assessment of importance. There is also an ambiguity in synonymy and polysemy, the connection between character strings and word senses. We argue that these limitations do not seriously impact the ability to detect relatedness in the corpus of biomedical research writing.
5.1 Context and focus
When a sentence is taken out of its surrounding context, some of the meaning of its implied references may be lost, e.g., anaphoric or omitted references. This loss of contextual meaning can make the judgment of relatedness ambiguous. In addition, whether or not two sentences are related in meaning can also depend on the relative importance that a particular reader places on the referenced subjects. As different readers may differ on what is important, they may also legitimately differ on the degree of relatedness of two sentences.
Here is an example that illustrates both limitations. In the manual evaluation, we encountered this pair of sentences:
-
(1)
Emissions were determined from field data by using a Fick’s law diffusion approach and the observed variation in time of the TCE concentration gradient within 4 m of each device (Wadden et al. 1989)
-
(2)
Observed and calculated fluxes based on vertical TCE vapor concentration gradients and Fick’s law were in good agreement (Jellali et al. 2003)
These two sentences both refer to emissions/fluxes of something that is observed based on “TCE concentration gradients” and calculated based on “Fick’s law.” To judge the degree of relatedness of these two sentences, one must consider both the emphasis and the missing context. Readers mainly interested only in TCE concentration gradients or Fick’s law, the techniques, might find these sentences to be very related. But, except for the reference to “devices” in the first sentence, the what, how, and why of the emissions, the subjects, are lost from the context. So readers mainly interested in the subjects would only find the sentences to be related if their missing subjects are the same or related.
In some cases it may be possible to infer the missing subjects from what is said about them. But in these examples, nearly identical things were said about the subjects. In fact, after reading the context of the sentences, we found that the first refers to industrial degreasing devices that use TCE (trichloroethylene), and the second refers to the environmental fate of TCE buried underground.
Fortunately, ambiguity of subject seemed to be an exception rather than the rule. There were only eight out of 1,000 sentences, like the above example, in which the evaluators gave opposite ratings (that is, 0 vs. 4). Most of the sentence pairs we observed contained explicit references to all of their important concepts. Such sentence pairs are ostensibly related because they refer to the same things, or unrelated because they refer to different things. But by using the average of the rating of two evaluators, we partially compensated for the ambiguous cases. For the above example, the average rating was 2, meaning possibly related, which seems to be about right.
5.2 Synonymy and polysemy
Theoretically, two sentences could assert very similar things using lexically unrelated synonyms. It was found, for example, that in task-oriented naming and keyword assignment, the same words were used to refer to the same things only 20% of the time (Furnas et al. 1987). To make matters worse, the technical vocabulary used in MEDLINE is significantly distinct and larger than that of newspapers and other casual media (Smith et al. 2005). But in scientific peer-reviewed publication, the requirement for clear communication encourages authors to use commonly accepted expressions when referring to any given concept. This is supported by the sublanguage hypothesis (Friedman et al. 2002), which holds that there are unique grammatical conventions that apply within a given scientific discipline. What is more, since a sublanguage also contains rules for the formation of new expressions, independent authors will tend to use lexically similar expressions, even when referencing a new concept.
In any event, it is unlikely that our corpus contained examples of lexically unrelated synonyms, since the positive examples were from the same abstract and the negative examples were from randomly paired sentences. Even so, our methods could recognize spelling and punctuation variants, and derived forms by comparing substring features. And sentences could still be identified as related even if some of their synonymous references were lexically distinct, provided that enough of their referenced concepts used common language.
Conversely, two sentences could refer to dissimilar things using lexically related homonyms, and this can occur in one of two ways (Bodenreider et al. 2002). In systematic polysemy, a word may be used to refer to distinct but closely related things. For example, genes and their products typically share the same name. This does not pose a problem for detecting relatedness, because all of the senses of such a word are related in meaning.
However, serious ambiguity occurs when a word may have entirely different meanings, depending on the context in which it is used, and this could result in false positives in relatedness detection. To explore how our system handled ambiguity, we examined sentences containing the ambiguous word ventilation. There were 36,357 sentences in the corpus which contained the word, and Table 5 shows four of the observed senses and the frequency of each sense in 1,000 randomly selected sentences. There were three sentences out of 1,000 that used ventilation in the sense of middle ear ventilation, and all three of their H-matches had the correct sense of the word. There were 12 sentences out of 1,000 that used ventilation in the sense of environmental flow. In four of those sentences, environmental air flow was not the dominant topic, and their corresponding H-matches did not refer to environmental air flow in any way. The H-matches for six of the remaining eight (75%) discuss environmental air flow, though only two actually contained the word ventilation. One of the erroneous H-matches used the respiration sense of the word, and one used the cigarette filter sense of the word. The following sentence pair illustrates a correct H-match on the environmental sense:
-
(1)
Among office workers, the relative risk for short-term sick leave was 1.53 (95% confidence 1.22–1.92) with lower ventilation, and 1.52 (1.18–1.97) in areas with IEQ complaints (Milton et al. 2000)
-
(2)
It is essential that treated areas be ventilated adequately before workers return to their offices (Currie et al. 1990)
While this match is not very strong (H-score of only 2.96) and not of high quality (the average of our ratings is 1.5), yet it illustrates how sufficient context can be included in a match (office, areas) to disambiguate the meaning of the word ventilation. In our sample we found that for the rare senses of the word ventilation, the H-match had the wrong sense of the word in only two of the cases we examined. In thirteen or 87% of the cases there is no confusion of meaning. Thus there is a strong tendency not to confuse the meaning of the word ventilation across the sentences of an H-match. While we cannot base general conclusions on such a small sample, we believe the principles acting here come into play more generally and provide at least a partial explanation for the success of our method.
6 Conclusion
We compared several approaches to identifying sentences in MEDLINE abstracts that are related in meaning based on a large automatically assembled training set. The baseline methods used fixed formulas that do not rely on training or optimization, and included the Dice coefficient, OKAPI bm25, the tf.idf cosine similarity used in TextTiling, and several idf-power functions. The machine learning methods included naïve Bayes and minimization of the Huber loss function. All of the methods achieved a BE greater than 84%. The differences between them were small but statistically significant, and favored the Huber loss function over the other methods.
Our ultimate goal was to find related sentence pairs coming from different abstracts. To achieve this goal, the best performing methods were applied to the problem of matching a query sentence with related sentences from other abstracts. When the top scoring matches were manually evaluated, we found a significant improvement of the Huber method (H) over both the Bayesian method (B) and the best baseline method (I 1.5), but no significant difference between the B and I 1.5 methods. When matches were chosen in the top 20% of Huber score (H ≥ 6.4), identifying about one sentence per abstract on average, there was an approximate precision of 73% and recall of 31%, as determined by manual evaluation (with positive relatedness defined by average human rating greater than 2). We have shown with an example (Fig. 4) how related sentences can be used to make focused suggestions for further reading.
In future work we plan to explore the use of our method of computing related sentences for topic segmentation and document summarization. There is also the possibility that it could be used to enhance full text retrieval.
Notes
With this definition, the cost \( C \) is scale invariant. That is to say, if \( w \) minimizes \( C \) with feature vectors \( x_{i} \) then for any \( \alpha \ne 0 \), \( \alpha^{ - 1} w \) minimizes \( C \) with feature vectors \( \alpha x_{i} \).
References
Artstein, R., & Poesio, M. (2008). Inter-coder agreement for computational linguistics. Computational Linguistics, 34(4), 555–596.
Bodenreider, O., Mitchell, J. A., & McCray, A. T. (2002). Evaluation of the UMLS as a terminology and knowledge resource for biomedical informatics. Proceedings of the AMIA symposium, pp. 61–65.
Currie, K. L., McDonald, E. C., Chung, L. T., & Higgs, A. R. (1990). Concentrations of diazinon, chlorpyrifos, and bendiocarb after application in offices. American Industrial Hygiene Association Journal, 51(1), 23–27.
Dietterich, T. (1998). Approximate statistical tests for comparing supervised classification learning algorithms. Neural Computation, 10(7), 1895–1924.
Friedman, C., Kra, P., & Rzhetsky, A. (2002). Two biomedical sublanguages: A description based on the theories of Zellig Harris. Biomedical Informatics, 35, 222–235.
Furnas, G. W., Landauer, T. K., Gomez, L. M., & Dumais, S. T. (1987). The vocabulary problem in human-system communication. Communications of the ACM, 30(11), 964–971.
Hearst, M. (1993). TextTiling: A quantitative approach to discourse. University of California at Berkeley. Report: S2 K-93-24.
Hearst, M., & Plaunt, C. (1993). Subtopic structuring for full-length document access. In: Proceedings of the ACM SIGIR conference, pp. 59–68.
Jellali, S., Benremita, H., Muntzer, P., Razakarisoa, O., & Schafer, G. (2003). A large-scale experiment on mass transfer of trichloroethylene from the unsaturated zone of a sandy aquifer to its interfaces. Journal of Contaminant Hydrology, 60(1–2), 31–53.
Joachims, T. (2002). Optimizing search engines using clickthrough data. In: Proceedings of the ACM SIGKDD conference, ACM, pp. 133–142.
Joachims, T. (2006). Training linear SVMs in linear time. In: Proceedings of the ACM SIGKDD conference, ACM, pp. 217–226.
Joachims, T., Li, H., Liu, T. -Y., & Zhai, C. (2007). Learning to rank for information retrieval (LR4IR 2007). In: Proceedings of the ACM SIGIR conference, ACM, pp. 58–62.
Kim, W. G., & Wilbur, W. J. (2001). Corpus-based statistical screening for content-bearing terms. Journal of the American Society for Information Science, 52(3), 247–259.
Ko, Y., Park, J., & Seo, J. (2002). Automatic text categorization using the importance of sentences. In: Proceedings of the 19th international conference on computational linguistics, pp. 65–79.
Langley, P. (1996). Elements of machine learning. San Francisco: Morgan Kaufmann Publishers, Inc.
Lin, J. (2009). Is searching full text more effective than searching abstracts? BMC Bioinformatics, 10, 46. doi:10.1186/1471-2105-10-46.
Lin, J., & Wilbur, W. (2007). PubMed related articles: A probabilistic topic-based model for content similarity. BMC Bioinformatics, 8 (423).
Liu, T. -Y., Xu, J., Qin, T., Xiong, W., & Li, H. (2007). Benchmark dataset for research on learning to rank for information retrieval. In: SIGIR ‘07: Proceedings of the Learning to Rank workshop.
Lu, Z., Kim, W., & Wilbur, W. (2009). Evaluating relevance ranking strategies for MEDLINE retrieval. Journal of the American Medical Informatics Association, 16(1), 32–36.
Ma, Q., Galasko, D., Ringman, J., Vinters, H., Edland, S., Pomakian, J., et al. (2009). Reduction of SorLA/LR11, a sorting protein limiting beta-amyloid production, in Alzheimer disease cerebrospinal fluid. Archives of Neurology, 66(4), 433–434.
Milton, D. K., Glencross, P. M., & Walters, M. D. (2000). Risk of sick leave associated with outdoor air supply rate, humidification, and occupant complaints. Indoor Air, 10(4), 212–221.
Robertson, S., & Spark Jones, K. (1994). Simple, proven approaches to text retrieval. University of Cambridge Computer Laboratory. Report: UCAM-CL-TR-356.
Salton, G., & Buckley, C. (1991). Automatic text structuring and retrieval-experiments in automatic encyclopedia searching. In: Conference on research and development in information retrieval, ACM, pp. 21–30.
Sayers, E. W., Barrett, T., Benson, D. A., Bryant, S. H., Canese, K., Chetvernin, V., et al. (2009). Database resources of the National Center for Biotechnology Information. Nucleic Acids Research, 37(Database Issue), 5–15.
Smith, L., Rindflesch, T., & Wilbur, W. (2004). MedPost: A part of speech tagger for biomedical text. Bioinformatics, 20(14), 2320–2321.
Smith, L., Rindflesch, T., & Wilbur, W. (2005). The importance of the lexicon in tagging biological text. Natural Language Engineering, 12(2), 1–17.
van Rijsbergen, C. J. (1979). Information retrieval. London: Butterworths.
Vapnik, V. (1998). Statistical learning theory. New York: Wiley.
Wadden, R., Scheff, P., & Franke, J. (1989). Emission factors for trichloroethylene vapor degreasers. American Industrial Hygiene Association Journal, 50(9), 496–500.
Wilbur, W. J. (1998). A comparison of group and individual performance among subject experts and untrained workers at the document retrieval task. Journal of the American Society for Information Science, 49(6), 517–529.
Wilbur, W. J. (2000). Boosting naive bayesian learning on a large subset of MEDLINE. In: American medical informatics 2000 annual symposium, American medical informatics association, pp. 918–922.
Wilbur, W. (2005). Modeling text retrieval in biomedicine. In H. Chen, S. S. Fuller, C. Friedman, & W. Hersh (Eds.), Medical informatics: knowledge management and data mining in biomedicine (pp. 277–297). New York: Springer.
Wilbur, W. J., & Sirotkin, K. (1992). The automatic identification of stop words. Journal of Information Science, 18, 45–55.
Zhang, T. (2004). Solving large scale linear prediction problems using stochastic gradient descent algorithms. In Proceedings of the Twenty-first international conference on machine learning, Omnipress, pp. 918–922.
Zou, H., Zhu, J., & Hastie, T. (2008). New multicategory boosting algorithms based on multicategory fisher-consistent losses. The Annals of Applied Statistics, 2(4), 1290–1306.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NLM, and NCBI.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Smith, L.H., Wilbur, W.J. Finding related sentence pairs in MEDLINE. Inf Retrieval 13, 601–617 (2010). https://doi.org/10.1007/s10791-010-9126-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10791-010-9126-8