Abstract
Diapausing egg banks are reservoirs of ecological and genetic diversity in continental zooplankton. However, although habitat size has often been used as a proxy for population size, the relationship between diapausing egg bank size and genetic diversity has not been explicitly tested in zooplankton. We estimated the density and size of diapausing egg banks, habitat size and genetic diversity (for mitochondrial and nuclear markers) of 14 populations of the rotifer Brachionus plicatilis in an endorheic basin in the SE Iberian Peninsula. The size of B. plicatilis diapausing egg banks ranged across eight orders of magnitude (from 257 to 4.9 × 1010 eggs). Despite the small geographical scale, populations were strongly structured genetically, but with no evidence of isolation by distance. Habitat size (lake area) was a better predictor of genetic diversity than total diapausing egg bank size, but only for mtDNA haplotype diversity. However, as these results were driven by the strong effect of the largest lake in the database, they should be taken with caution. Our results suggest that large lakes could have a disproportionate effect on genetic diversity and that more work is needed to support the use of habitat size as a proxy of population size in rotifers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most groups of continental zooplankton produce egg banks, accumulations of large numbers of diapausing eggs analogous to annual plant seed banks (Brendonck & De Meester, 2003). Diapausing egg banks are reservoirs of both neutral and adaptive genetic variation, and function as a population memory of past selective conditions (Cousyn et al., 2001). Of crucial importance, they allow population survival during adverse periods and can facilitate species coexistence (Cáceres, 1997; Montero-Pau et al., 2011). Largely due to these features, and the fact that diapausing egg banks are present in habitats year round, diapausing egg banks have been used to investigate zooplankton phylogeography (Gómez et al., 2000a, 2007), the distribution of cryptic species (Ortells et al., 2000), community diversity (May, 1987), cryptic species coexistence (Montero-Pau et al., 2011), and species invasions (Mergeay et al., 2006).
Population genetics theory predicts an effect of population size on genetic diversity, which has been subject of much research (for a recent assessment, see Leffler et al., 2012). A positive effect of population size on genetic diversity is expected because smaller populations suffer from higher genetic drift, and therefore, they will lose diversity at a higher rate (Falconer, 1989). Large populations are also expected to be inherently more stable, subject to lower demographic and environmental stochasticity, and reduced extinction rates (Lande, 1988; Frankham, 2005). Thus, from a metapopulation perspective approach, large populations could serve as propagule sources for small populations, which in turn would act as genetic sinks more prone to extinction/recolonization cycles. Both genetic and demographic effects should contribute to a positive association between population size and genetic diversity. In metapopulations of species of the zooplanktonic crustacean genus Daphnia, younger populations, which are affected by strong founder effects and inbreeding depression and are more prone to extinction, are also genetically less diverse than older ones, which act as reservoirs or sources of genetic diversity for the smaller populations (Ebert et al., 2002; Ilmari Pajunen & Pajunen, 2003; Haag et al., 2005). Inbreeding depression has also been documented in laboratory rotifer populations (Tortajada et al., 2009). However, the relationship between population size and genetic diversity has not been explicitly tested in zooplankton.
Both the estimation of population sizes themselves and genetic diversity in zooplankton is problematic using active populations in the water column, as they are strongly fluctuating at a seasonal, annual, and longer time scales (Gómez et al., 1995; Montero-Pau & Gómez, 2011). In addition, the use of populations from the water column or hatched from diapausing eggs suffers from the problem of clonal reproduction or biased hatching; these will underestimate the genetic diversity of a population (Gómez et al., 2002a). Therefore, diapausing egg bank size, which is inherently more stable and produced sexually, could potentially be a good proxy for effective population size. Another potentially useful proxy for effective population size is habitat size (Hänfling & Brandl, 1998). The size of a habitat might be highly correlated with the maximum population size that can be attained and therefore limit its genetic diversity (Keyghobadi, 2007). Indeed, previous studies in the planktonic crustaceans Daphnia magna Straus and Artemia sinica Cai have investigated the relationship between estimates of zooplankton habitat size (lake or pond volume or area) and genetic diversity estimated from the active population. In highly dynamic D. magna metapopulations in Northern Europe, pond volume is significantly associated to both population age and genetic diversity estimated from the population active in the water column (Haag et al., 2005). In the same species, northern—younger, dwelling in small ponds—populations had lower genetic diversity for both mtDNA and microsatellites than southern—older, dwelling in large ponds—populations (Walser & Haag, 2012). Although historical effects (e.g., postglacial recolonization processes) could potentially explain that northern populations have lower diversity, such effects are less likely to operate on microsatellite diversity due to the higher mutation rates in these markers. Also in D. magna, pond area and genetic and clonal diversity were significantly correlated in 17 ponds in Belgium using four allozyme loci (Vanoverbeke et al., 2007). In Artemia sinica populations, genetic diversity, as estimated from allozymes, was correlated with habitat size (Naihong et al., 2000). As the study included very distant populations of unknown phylogeographic history (max distance 1200 km), historical factors might potentially confound the observed correlation (Walser & Haag, 2012). In these studies, however, higher genetic diversities could reflect either higher population sizes or higher habitat stability in larger habitats, the latter being due to either at ecological (e.g., lower environmental fluctuations in larger habitats) or historical factors (e.g., larger lakes being refuges). In addition, differences in habitat quality unrelated to habitat size (e.g., salinity differences and degree of eutrophication), or the presence of competing species might affect population sizes, and therefore, habitat size per se might not be a good proxy for population size.
Brachionus plicatilis Müller, 1786, hereafter B. plicatilis sensu stricto (s. s.), is a rotifer belonging to a complex of cryptic species found in salt lakes worldwide (Gómez et al., 2002b; Suatoni et al., 2006; Mills et al., 2007), with species often co-occurring in the same lakes (Ortells et al., 2003; Gómez, 2005; Montero-Pau et al., 2011). Populations are typically seasonal and reproduce through cyclical parthenogenesis, with the first generation of clonally reproducing females hatching from sexually produced diapausing eggs. After a variable number of generations of clonal reproduction, a sexual generation occurs, which produce diapausing eggs. Diapausing eggs are long lived and accumulate in large numbers forming relatively shallow and young egg banks in the habitat sediments (Ortells et al., 2000; García-Roger et al., 2006a, b) where they persist through unfavorable periods. B. plicatilis s. s. shows strong population and phylogeographic structure, with evidence of an ‘isolation by distance’ pattern at the regional and continental scales, likely as a result of sequential colonization (Gómez et al., 2000b, 2002a; Mills et al., 2007; Campillo et al., 2011). In many areas in Eastern Spain, particularly in inland salt lakes, B. plicatilis s. s. and another species in the species complex, B. manjavacas Fontaneto et al., 2007, which compete due to their strong niche overlap (Gabaldón et al., 2013), often co-occur (Gómez, 2005).
Here we explore the relationship between the size and density of diapausing egg banks, habitat size, and genetic diversity in populations of the rotifer B. plicatilis s. s. in salt lakes from a small endorheic basin in the SE Iberian Peninsula. Previous phylogeographic analysis suggests that populations in this basin are quite homogeneous genetically and therefore likely to have been founded in a similar time scale (Gómez et al., 2000a; Campillo et al., 2011). We use both mitochondrial and nuclear microsatellite markers, the former more likely to have stronger genetic drift (Ballard & Whitlock, 2004).
Methods
Study area and diapausing egg isolation
We intensively sampled an endorheic area of about 275 km2 containing salt lakes in the Júcar-Segura basin (Albacete province, Spain) where B. plicatilis s. s. is common (Gómez et al., 2000a). Using cartographic maps, we identified 34 potential sampling points (lakes, ponds, and pools; Fig. 1), which we visited between October 2004 and January 2005. Sediment samples were taken either if salinity suggested the occurrence of these species or, if the basin was dry, the place showed no signs of being cultivated, resulting in 19 lakes being sampled. As diapausing egg banks in Brachionus rotifers are very shallow, with most viable diapausing eggs concentrated in the top centimeters of the sediment (García-Roger et al., 2006a), we collected two superficial sediment samples per lake using a scoop, one in the deepest point and another in the shallowest, in order to integrate the maximum possible environmental variability. In three occasions (lakes coded as HYM, SLD and PET; Table 1), the lake center could not be reached, hence samples were taken from two distant point on the shores. Samples were stored in dark and cool conditions until required. Additionally, Pétrola lake (PET) was sampled in December 2005–October 2006 as part of a parallel study (Montero-Pau et al., 2011) using sediment cores. Thus, for Pétrola population, diapausing egg bank density (eggs g−1) and genetic data of 16S rRNA in Montero-Pau et al. (2011) were included in this study’s dataset for analysis, whereas data for mitochondrial cytochrome c oxidase subunit I (COI) and microsatellites were newly obtained in this study. In order to achieve larger sample sizes, given that a competing species was dominant on the superficial sediment of this lake; data from the first cm of cores 1 and 2 (see Montero-Pau et al., 2011) were grouped as Pétrola North Shore and data from cores 3 and 4, as Pétrola South Shore, respectively.
Diapausing eggs were isolated using a sugar flotation technique (Onbé, 1978; Gómez & Carvalho, 2000) from 53 to 736 g of dry sediment until either at least 200 eggs that could be morphologically assigned to the Brachionus plicatilis species complex had been isolated; or after processing 200 g, a total of less than 10 eggs were found. Eggs were counted under a stereomicroscope, and diapausing egg density per gram of superficial sediment processed was then calculated for the complex (hereafter, total diapausing egg density). Egg density (egg g−1) was transformed into egg density per cm3 using the value of sediment density in Pétrola lake (1.56 g cm−3) (JMP, personal observation) and then to egg density per cm2. As lake area is strongly fluctuating, we used maximum lake area estimated from Google Earth’s satellite images using ImageJ v.1.48 (Schneider et al., 2012) (Suppl. Fig. 1). Lake area is best estimated with satellite imagery as the maximum lake shore can be easily recognized by the fringe of reed beds and other shore vegetation. Egg bank size of the complex (hereafter, total egg bank size) was estimated by multiplying maximum lake area by average egg density of the two sampled points, and thus assuming that the two sampling points are representative of equivalent areas of the lake. B. plicatilis s. s. eggs cannot be morphologically distinguished from those of other species in its species complex. Thus species identification was based on the analysis of a fragment of the mitochondrial gene 16S rRNA (Montero-Pau et al., 2011), see below). With this information, we estimated (i) the frequency of viable diapausing eggs for B. plicatilis s. s., (ii) the presence, and frequency of competing species from the species complex in the diapausing egg bank, and (iii) the egg density and egg bank size of B. plicatilis s. s.
SSCP analysis of mitochondrial genes and species identification
For each individual egg isolated from the sediment, we amplified a 378 bp fragment of the mitochondrial gene 16S rRNA using the primers described in Papakostas et al. (2005). DNA was extracted from single diapausing eggs using a modified alkaline lysis protocol (HotSHOT) (Montero-Pau et al., 2008). We used PCR-single-stranded conformation polymorphism (SSCP) analysis as described in Montero-Pau et al. (2011). Samples were assigned to an electromorph pattern by eye, and one sample from each electromorph per gel was sequenced. PCR amplifications were repeated for samples selected for sequencing under the same conditions but in a final volume of 50 µl. Products were purified using High Pure PCR Product Purification Kit (Roche) and sequenced using ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Biosystems) in both directions and run in an ABI 3700 sequencer (Perkin-Elmer Biosystems). Chromatograms were checked and edited using CodonCode Aligner v. 1.6. (CodonCode Corporation). Samples presenting the same electromorph were then assigned the same species and haplotype than the corresponding sequenced sample. For species identification (barcoding), in order to screen B. plicatilis s. s. samples for microsatellites, and to estimate the number of diapausing eggs in the sediment samples for each species, we used the results obtained for the 16S fragment, after Papakostas et al. (2005), and Montero-Pau et al. (2011). Individuals identified as B. plicatilis s. s. were also genotyped for a 323 bp fragment of COI using PCR-SSCP following the same procedure as above, amplified with primers LCO1490 (Folmer et al., 1994) and BrCoi301L25 (5′-AGGACAACAGAAGAAACATAAAGGC-3′), which was designed for this study.
Microsatellite genotyping
Seven polymorphic microsatellite loci (Gómez et al., 1998) were amplified for individual diapausing eggs identified as B. plicatilis s. s. from the results of the 16S SSCP analysis. We followed PCR conditions in Gómez et al. (1998). PCR products were multiplexed and separated in a Beckman-Coulter TM CEQ 8000 Genome Analysis System. Peaks were scored by eye using the CEQ Fragment Analysis software (Beckman–Coulter TM). Microchecker v. 2.2 (Van Oosterhout et al., 2004) was used to check for the presence of null alleles, scoring errors, or evidence of allele dropout.
Genetic diversity
Unbiased genetic diversity, number of alleles, and Shannon–Weaver diversity index for each population were estimated for microsatellite and mitochondrial data using GenAlEx v. 6.501 (Peakall & Smouse, 2012). For microsatellites, observed heterozygosity and fixation index were also estimated. For mitochondrial data, indexes were calculated for loci 16S and COI as well as for the concatenated data. 95% confidence intervals for genetic diversity were obtained using bootstrap resampling and corrected following the bias-corrected percentile method (Efron, 1981) using R v.3.1.3 (R Core Team, 2015) with a total of 1,000 randomizations for each sampling point. To account for different sampling sizes, a rarefaction analysis was performed on the number of alleles and haplotypes using function allelic.richness in R package ‘hierfstat’ (Goudet, 2005). Only populations with nine or more genotyped individuals were used. A paired t test was used to compare genetic diversity of microsatellites and mitochondrial genes between center/shore sampling points using the R software.
Population genetic analysis
Tests for deviations from Hardy–Weinberg and linkage disequilibrium were carried out using Genepop v. 4.0. (Raymond & Rousset, 1995) Global and population pairwise genetic differentiation were computed using Arlequin (Excoffier et al., 2005); Φst was estimated for mtDNA data and F st values were obtained for microsatellites. Statistical significance of genetic differentiation was assessed by performing 10,000 permutations. An Analysis of Molecular Variance (AMOVA) as implemented in Arlequin was also performed for mitochondrial and microsatellite genes with sampling point and lake as factors. A non-metric multidimensional scaling (NMDS) was performed based on genetic distances (Φst and F st) using metaMDS function in R package ‘vegan’ (Oksanen et al., 2015). Patterns of isolation by distance for microsatellites and mtDNA was assessed using Mantel tests using mantel.rtest function in the R package ‘ade4’ (Dray et al., 2007).
Correlation between genetic diversity and proxies of population size
Correlations were assessed in order to analyze linear relationships between population size proxies (lake area, B. plicatilis s. s. diapausing egg density, size B. plicatilis s. s. egg bank and proportion of B. plicatilis s. s.) and genetic diversity measurements (dependent variables, measured using expected heterozygosity and number of alleles/haplotypes corrected by rarefaction). Correlations were also performed using the logarithm of lake area and size of B. plicatilis s. s. egg bank in order to reduce the effect of the skewed distribution of these two parameters. Additionally, a partial least squares path modeling analysis was performed using plspm function in R package ‘plspm’ (Sanchez et al., 2013). This analysis assumes a model with paths (unidirectional or bidirectional relationships) defined a priori and estimates the direct and indirect effect of latent variables based on partial regression coefficients between measured variables. We established a model with three latent variables (lake area, population size, and genetic diversity). Lake area was measured as area of lake, population size as size of B. plicatilis s. s. diapausing egg bank and density of B. plicatilis s. s. diapausing egg, and genetic diversity as unbiased genetic diversity and rarefaction-corrected number of alleles/haplotypes for microsatellite and mtDNA data, respectively.
Results
Diapausing egg bank densities
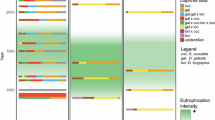
Sediment samples were obtained from 19 lakes, of which eight were dry at the time of sampling. Brachionus complex diapausing eggs were found in 14 of the lakes sampled (Table 1; Fig. 2). Species identification of the specimens of the species complex based on 16S haplotypes shows that B. plicatilis s. s. was found in all 14 lakes, while B. manjavacas, the next more common species, was found in seven lakes, mainly in the central samples, with 2–56% of the total eggs analyzed (Tables 1, 2). B. ‘almenara’ was found in Ontalafia and Pétrola lake and B. ibericus Ciros-Pérez et al., 2001 was only found in Ontalafia. The estimated density of B. plicatilis s. s. eggs ranged from 0.003 in HYC to 2.154 eggs g−1 in HYB lake. Estimated B. plicatilis s. s. egg bank size varied in eight orders of magnitude, ranging from 257 to 4.9 × 1010 eggs per lake (Fig. 2). Although total diapausing eggs density and B. plicatilis s. s. eggs density tended to be higher in the center (Table 1), differences were not statistically significant (paired t test: t = 0.643, df = 10, P value = 0.535, and t = 1.093, df = 10, P value = 0.300 respectively). In the four lakes, where eggs were found in both shore and center and B. plicatilis s. s. co-occurred with other species of the complex, B. plicatilis s. s. was found in higher frequency on the shore than in the center in three of them, but this difference was significant only in one lake (SAL; based on 95% binomial confidence interval) (Suppl. Fig. 2).
Diapausing egg banks for the Brachionus plicatilis species complex in the studied lakes. Bars show egg density for each species in the complex (dark gray: B. plicatilis s. s.; light gray: B. manjavacas; black: B. ibericus and B. almenara). Dots are egg bank size of B. plicatilis s. s. in each lake as estimated from maximum lake area. Lakes are ordered by increasing maximum area
We found the expected positive correlation between total egg density for the species complex and lake area (R 2 = 0.81, P value one-sided hypothesis <0.001). However, no correlation was found between lake area and B. plicatilis s. s. egg density (R 2 = 0.21, P value = 0.103).
Mitochondrial DNA diversity
We extracted and analyzed a total of 665 diapausing eggs for 16S and 549 for COI. To this, we added data from gene 16S from 347 diapausing eggs of the species complex from Pétrola obtained in a previous study (Montero-Pau et al., 2011), and we genotyped the same eggs additionally for COI in B. plicatilis s. s. A total of four haplotypes for 16S and five for COI were found in B. plicatilis s. s. The concatenation of both mitochondrial fragments resulted in a total of eight B. plicatilis s. s. haplotypes, three of them only found in Pétrola lake, one only found in Salobrejo, and another only in Hoya del Monte (Table 2, Suppl. Tables 1, 2; Fig. 3). Pétrola lake had the highest number of haplotypes, seven, and highest haplotype diversity (Table 2), whereas HTU, despite a large sample size (40), had a single haplotype.
Microsatellite diversity
We found evidence of either null alleles or scoring errors in HYR, ATA, SLD, and PET for microsatellite Bp3 (raw data in Suppl. Table 3). Therefore, this locus was excluded from further analysis. Samples sizes, allelic diversity, and expected and observed heterozygosity are shown on Table 2 and Suppl. Table 1. Loci were polymorphic in most populations, although locus Bp4a that was only polymorphic in HYB, HYR, and ONT, and loci Bp1, Bp5, and Bp6 were not variable in HTU and SLD, SAL and HTU, respectively. Mean expected heterozygosity was highest in ONT (0.475) and lowest in SAL (0.252). Statistically significant evidence for linkage disequilibrium was found between loci Bp2 and Bp3c in ONT, Bp3c and Bp5 in HYM, and Bp2 and Bp6 in SLD (data not shown). No evidence of H-W disequilibrium was found for any lake, either if sampling points within lake were considered separately or if they were merged.
Population structure and isolation by distance
Populations with less than nine diapausing eggs of B. plicatilis s. s. genotyped (HYC, HYE, MJB, and MSA) were not used in analyses of population structure and isolation by distance. Population genetic structure was very high (Fig. 4), with pairwise F st for concatenated mitochondrial genes ranging from 0 to 0.762 and global F st was 0.477 (Table 3). According to MANOVA, the variance explained by the differentiation between sampling points (shore, center) is 1.69%, while the variance explained among lakes is 46.04%. For microsatellites, population differentiation ranged from 0.054 to 0.584 and global F st was 0.253. The variance explained by the differentiation among sampling points is 1.04%, while the variance explained among lakes is 24.30%.
No significant differences were found for the genetic diversity between shore/center sampling points (paired t test: t = 1.703, df = 3, P value = 0.187 for microsatellites; t = 1.430, df = 3, P value = 0.248 for the concatenation of mitochondrial genes) (Suppl. Fig. 3). No evidence of isolation by distance was found (Suppl. Fig. 4) neither for the mitochondrial genes nor microsatellites.
Relationships between B. plicatilis s. s. diapausing bank size, lake area, and genetic diversity
For microsatellites, no significant correlation was found between genetic diversity (using either expected heterozygosity, He, or number of alleles corrected by rarefaction, rNa) and B. plicatilis s. s. diapausing egg bank size or lake size (Fig. 5; Suppl. Table 4). In contrast, for mtDNA, significant correlations were found between the rarefaction-corrected number of haplotypes (rNa) on the one hand and lake area and B. plicatilis s. s. diapausing egg bank size, on the other hand (Fig. 5). As lake area and size of B. plicatilis s. s. diapausing egg bank have a skewed distribution, correlations were also assessed using the log-transformation of both variables to reduce this effect. When using log-transformed variables, statistical significance only was found between rNa and lake area (Fig. 5). Pétrola, the largest lake by far, and also the lake that held the largest B. plicatilis s. s. diapausing egg bank, also has the highest heterozygosity and allelic diversity. To assess the effect of this potential outlier in our results, we repeated the analyses removing Pétrola from the dataset no significant effects were found (Suppl. Fig. 5).
Relationship between the number of alleles/haplotypes corrected by rarefaction and B. plicatilis s. s. egg bank size (two superior panels) and lake size (two inferior panels) for microsatellites (left panels) and mitochondrial concatenate (right panels). Lines were fitted using least squares linear regression using untransformed (dashed line) or log-transformed (solid line) population size proxies
The partial least squares path modeling analysis of the relationship among latent variables (lake area, population size, and mitochondrial genetic diversity) revealed that lake area had a highly significant effect on B. plicatilis s. s. population size (direct path coefficient = 0.891; P value <0.001; Fig. 6), although not on genetic diversity (direct path coefficient = 0.265; P value = 0.626). Population size did not have a direct significant effect on rNa (direct path coefficient = 0.535; P value = 0.338). Altogether, lake area resulted in a strong total effect of lake area on rNA (indirect path coefficient = 0.265, total path coefficient = 0.744). However, after excluding Pétrola, the explanatory power of the model weakened (R 2 dropped from 0.764 to 0.442), and none of the direct effects were significant (area on population size: direct path coefficient = 0.638, P value = 0.064; area on genetic diversity: direct path coefficient = −0.151, P value = 0.772; population size on genetic diversity: direct path coefficient = 0.423, P value = 0.428). Also, total effect (direct and indirect) of area on rNa dropped from 0.744 to 0.119.
Discussion
Our results suggest that although lake area is a better predictor of population genetic diversity than the size of B. plicatilis s. s. diapausing egg banks, this effect was only significant when population genetic diversity was estimated for mtDNA, but not for nuclear DNA, and was due to a disproportionate effect of the largest lake Pétrola.
Diapausing egg bank densities, lake area, and competitive interactions
Superficial diapausing egg bank densities of B. plicatilis s. s. and the absolute sizes of their egg banks showed an enormous large range of variation (i.e., eight orders of magnitude for total diapausing eggs banks) and reached comparable sizes to those estimated for other zooplanktonic organisms (De Meester et al., 2004). The fact that larger lakes hold higher diapausing egg bank densities of the species complex suggest that they might be more ecologically suitable for these rotifers, possibly through higher environmental stability. However, we did not find an effect of lake size on B. plicatilis s. s. egg density, what may reflect its competitive relationships with B. manjavacas, a common species in this group of lakes with similar ecological requirements (Gabaldón et al., 2013). These competitive relationships result on strong fluctuations, with an alternation in dominance between these two species reflected in their diapausing egg banks (Montero-Pau et al., 2011). Indeed, the trend of B. plicatilis s. s. to have higher density on the shore when coexisting with B. manjavacas might be reflecting dominance of B. plicatilis s. s. in episodes of lake filling associated to lower salinity and reflecting B. manjavacas better adaptation to higher salinity (Gabaldón et al., 2015).
Population structure and isolation by distance
As previously observed in rotifer populations at larger geographical scales (Gómez et al., 2000b; Mills et al., 2007), population structure was remarkable, with high genetic differentiation between lakes and global F st of 0.477 for mtDNA. Similarly, for microsatellites, population differentiation was high, although not as much as for mtDNA with global F st was 0.239. We found no evidence of an ‘isolation by distance’ pattern neither for the mitochondrial genes nor for microsatellites, in contrast to previous regional or global assessments (Gómez et al., 2002a; Mills et al., 2007). Sequential colonization effect might work at larger scales (regional/continental), but it is less likely to work at local scales, where a highly dynamic process of extinction and recolonization might be dominant.
Population size and genetic diversity
We found no effect of proxies of population size on microsatellite diversity, contrasting with significant effects of two proxies of population size (lake area and diapausing egg bank size) on mtDNA number of haplotypes. As the effective population size of mtDNA markers is four times smaller than in nuclear DNA markers, mtDNA suffers from stronger genetic drift, and therefore smaller lakes holding small populations might lose mtDNA diversity faster than nuclear DNA (Ballard & Whitlock, 2004). Given the effort involved in estimating diapausing egg banks, our results suggest that lake area, which is easily estimated from maps or aerial imagery, could be the a very cost-effective estimator of population size, especially if there are strong time fluctuations in the size of the diapausing egg bank. This has been shown to be the case, for example, when two competing species might strongly alternate in abundance due to changing ecological conditions, like in the case of B. plicatilis s. s. and B. manjavacas in Pétrola (Montero-Pau et al., 2011). Therefore, competitive processes resulting in strong interannual species fluctuations will reduce the usefulness of superficial diapausing egg bank sizes as proxy of effective population sizes and therefore weaken any correlation with genetic diversity.
We must be cautious, however, in not overstating our results as the statistical significance of the relationship between mtDNA and proxies for population size relies on the large effect of Pétrola, the largest lake in the dataset. Unfortunately, in the sampled system, there was no additional pond similar in size to Pétrola, so that a caveat exists on whether the high genetic diversity harbored in Pétrola is due to its size or to an unknown different singularity. Nevertheless, the ponds included in the study are in a small geologically and geomorphologically very homogeneous area, so that pond differences not due to size are unlikely factors affecting to genetic diversity. By contrast, it is possible that large lakes have a disproportionate effect on genetic diversity, mediated by their larger population sizes, and lower ecological stochasticity, but the confirmation of this effect will require further study. The generality of our results should be assessed by including more lakes and investigating the relationship between genetic diversity and lake area in other zooplanktonic organisms. In addition, in order to resolve the effects of large population sizes and ecological stochasticity on genetic diversity, estimates of population ages, possibly based on paleolimnological studies, would be very informative.
References
Ballard, J. W. O. & M. C. Whitlock, 2004. The incomplete natural history of mitochondria. Molecular Ecology 13: 729–744.
Brendonck, L. & L. De Meester, 2003. Egg banks in freshwater zooplankton: evolutionary and ecological archives in the sediment. Hydrobiologia 491: 65–84.
Cáceres, C. E., 1997. Temporal variation, dormancy, and coexistence: a field test of the storage effect. Proceedings of the National Academy of Sciences of the United States of America 94: 9171–9175.
Campillo, S., M. Serra, M. J. Carmona & A. Gómez, 2011. Widespread secondary contact and new glacial refugia in the halophilic rotifer Brachionus plicatilis in the Iberian Peninsula. PloS One 6: e20986.
Cousyn, C., L. De Meester, J. K. Colbourne, L. Brendonck, D. Verschuren & F. Volckaert, 2001. Rapid, local adaptation of zooplankton behavior to changes in predation pressure in the absence of neutral genetic changes. Proceedings of the National Academy of Sciences of the United States of America 98: 6256–6260.
De Meester, L., A. Gomez & J. C. Simon, 2004. Evolutionary and Ecological Genetics of Cyclical Parthenogens Evolution: From Molecules to Ecosystems. Oxford University Press, Oxford.
Dray, S., A.-B. Dufour, & Others, 2007. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22: 1–20.
Ebert, D., C. Haag, M. Kirkpatrick, M. Riek, J. W. Hottinger & V. I. Pajunen, 2002. A selective advantage to immigrant genes in a Daphnia metapopulation. Science 295: 485–488.
Efron, B., 1981. nonparametric standard errors and confidence intervals. The Canadian Journal of Statistics 9: 139–158.
Excoffier, L., G. Laval & S. Schneider, 2005. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1: 47–50.
Falconer, D. S., 1989. Introduction to Quantitative Genetics. Longman, Harlow.
Folmer, O., M. Black, W. Hoeh, R. Lutz & R. Vrijenhoek, 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology 3: 294–299.
Frankham, R., 2005. Genetics and extinction. Biological Conservation 126: 131–140.
Gabaldón, C., J. Montero-Pau, M. Serra & M. J. Carmona, 2013. Morphological similarity and ecological overlap in two rotifer species. PloS One 8: e57087.
Gabaldón, C., M. Serra, M. J. Carmona & J. Montero-Pau, 2015. Life-history traits, abiotic environment and coexistence: the case of two cryptic rotifer species. Journal of Experimental Marine Biology and Ecology 465: 142–152.
García-Roger, E. M., M. J. Carmona & M. Serra, 2006a. Hatching and viability of rotifer diapausing eggs collected from pond sediments. Freshwater Biology 51: 1351–1358.
García-Roger, E. M., M. J. Carmona & M. Serra, 2006b. Patterns in rotifer diapausing egg banks: density and viability. Journal of Experimental Marine Biology and Ecology 336: 198–210.
Gómez, A., 2005. Molecular ecology of rotifers: from population differentiation to speciation. Hydrobiologia 546: 83–99.
Gómez, A. & G. R. Carvalho, 2000. Sex, parthenogenesis and genetic structure of rotifers: microsatellite analysis of contemporary and resting egg bank populations. Molecular Ecology 9: 203–214.
Gómez, A., M. Temprano & M. Serra, 1995. Ecological genetics of a cyclical parthenogen in temporary habitats. Journal of Evolutionary Biology 8: 601–622.
Gómez, A., C. Clabby & G. R. Carvalho, 1998. Isolation and characterization of microsatellite loci in a cyclically parthenogenetic rotifer, Brachionus plicatilis. Molecular Ecology 7: 1619–1621.
Gómez, A., G. R. Carvalho & D. H. Lunt, 2000a. Phylogeography and regional endemism of a passively dispersing zooplankter: mitochondrial DNA variation in rotifer resting egg banks. Proceedings of the Royal Society of London, Series B 267: 2189–2197.
Gómez, A., G. R. Carvalho & D. H. Lunt, 2000b. Phylogeography and regional endemism of a passively dispersing zooplankter: mitochondrial DNA variation in rotifer resting egg banks. Proceedings of the Royal Society of London, Series B 267: 2189.
Gómez, A., G. J. Adcock, D. H. Lunt & G. R. Carvalho, 2002a. The interplay between colonization history and gene flow in passively dispersing zooplankton: microsatellite analysis of rotifer resting egg banks. Journal of Evolutionary Biology 15: 158–171.
Gómez, A., M. Serra, G. R. Carvalho & D. H. Lunt, 2002b. Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution 56: 1431–1444.
Gómez, A., J. Montero-Pau, D. H. Lunt, M. Serra & S. Campillo, 2007. Persistent genetic signatures of colonization in Brachionus manjavacas rotifers in the Iberian Peninsula. Molecular Ecology 16: 3228–3240.
Goudet, J., 2005. Hierfstat, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes 5: 184–186.
Haag, C. R., M. Riek, J. W. Hottinger, V. I. Pajunen & D. Ebert, 2005. Genetic diversity and genetic differentiation in Daphnia metapopulations with subpopulations of known age. Genetics 170: 1809–1820.
Hänfling, B. & R. Brandl, 1998. Genetic variability, population size and isolation of distinct populations in the freshwater fish Cottus gobio L. Molecular Ecology 7: 1625–1632.
Ilmari Pajunen, V. & I. Pajunen, 2003. Long-term dynamics in rock pool Daphnia metapopulations. Ecography 26: 731–738.
Keyghobadi, N., 2007. The genetic implications of habitat fragmentation for animals. Canadian Journal of Zoology 85: 1049–1064.
Lande, R., 1988. Genetics and demography in biological conservation. Science 241: 1455–1460.
Leffler, E. M., K. Bullaughey, D. R. Matute, W. K. Meyer, L. Ségurel, A. Venkat, P. Andolfatto & M. Przeworski, 2012. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biology 10: e1001388.
May, L., 1987. Effect of incubation temperature on the hatching of rotifer resting eggs collected from sediments. Hydrobiologia 147: 335–338.
Mergeay, J., D. Verschuren & L. De Meester, 2006. Invasion of an asexual American water flea clone throughout Africa and rapid displacement of a native sibling species. Proceedings of the Royal Society of London, Series B 273: 2839–2844.
Mills, S., D. H. Lunt & A. Gómez, 2007. Global isolation by distance despite strong regional phylogeography in a small metazoan. BMC Evolutionary Biology 7: 225.
Montero-Pau, J. & A. Gómez, 2011. Development of genomic resources for the phylogenetic analysis of the Brachionus plicatilis species complex (Rotifera: Monogononta). Hydrobiologia 662: 43–50.
Montero-Pau, J., J. Muñoz & A. Gómez, 2008. Application of an inexpensive and high-throughput genomic DNA extraction method for the molecular ecology of zooplanktonic diapausing eggs. Limnology and Oceanography, Methods 6: 218–222.
Montero-Pau, J., E. Ramos-Rodríguez, M. Serra & A. Gómez, 2011. Long-term coexistence of rotifer cryptic species. PloS One 6: e21530.
Naihong, X., E. Audenaert, J. Vanoverbeke, L. Brendonck, P. Sorgeloos & L. De Meester, 2000. Low among-population genetic differentiation in Chinese bisexual Artemia populations. Heredity 84: 238–243.
Oksanen, J., F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, R. B. O’Hara, G. L. Simpson, P. Solymos, M. H. H. Stevens, H. Wagner, & Others, 2015. Package “vegan.” Community ecology package, version 2.2.
Onbé, T., 1978. Sugar flotation method for sorting the resting eggs of marine cladocerans and copepods from sea-bottom sediment. Bulletin of the Japanese Society of Scientific Fisheries 44: 1411–1412.
Ortells, R., A. Gómez & M. Serra, 2003. Coexistence of cryptic rotifer species: ecological and genetic characterisation of Brachionus plicatilis. Freshwater Biology 48: 2194–2202.
Ortells, R., T. W. Snell, A. Gomez & M. Serra, 2000. Patterns of genetic differentiation in resting egg banks of a rotifer species complex in Spain. Archiv für Hydrobiologie 149: 529–551.
Papakostas, S., A. Triantafyllidis, I. Kappas & T. J. Abatzopoulos, 2005. The utility of the 16S gene in investigating cryptic speciation within the Brachionus plicatilis species complex. Marine Biology 147: 1129–1139.
Peakall, R. & P. E. Smouse, 2012. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28: 2537–2539.
R Core Team, 2015. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna.
Raymond, M. & F. Rousset, 1995. GENEPOP (Version 1.2): population genetics software for exact tests and ecumenicism. The Journal of Heredity 86: 248–249.
Sanchez, G., L. Trinchera, & G. Russolillo, 2013. plspm: tools for partial least squares path modeling (PLS-PM). R package version 0.4.1.
Schneider, C. A., W. S. Rasband & K. W. Eliceiri, 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9: 671–675.
Suatoni, E., S. Vicario, S. Rice, T. W. Snell & A. Caccone, 2006. Phylogenetic and biogeographic patterns in the salt water rotifer Brachionus plicatilis. Molecular Phylogenetics and Evolution 41: 86–98.
Tortajada, A. M., M. J. Carmona & M. Serra, 2009. Does haplodiploidy purge inbreeding depression in rotifer populations? PloS One 4: e8195.
Van Oosterhout, C., W. F. Hutchinson, D. P. M. Wills & P. Shipley, 2004. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4: 535–538.
Vanoverbeke, J., K. De Gelas & L. De Meester, 2007. Habitat size and the genetic structure of a cyclical parthenogen, Daphnia magna. Heredity 98: 419–426.
Walser, B. & C. R. Haag, 2012. Strong intraspecific variation in genetic diversity and genetic differentiation in Daphnia magna: the effects of population turnover and population size. Molecular Ecology 21: 851–861.
Acknowledgments
This work was funded by a grant from the Spanish Ministerio de Ciencia e Innovación (CGL2009-07364) to MS. AG was supported by a National Environment Research Council (NERC) Advanced Fellowship (NE/B501298/1) and the University of Hull and JM-P by a fellowship by the Spanish Ministerio de Ciencia y Tecnología (BES2004-5248). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We are grateful to Lluís Franch for constructive comments on previous versions of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: M. Devetter, D. Fontaneto, C. D. Jersabek, D. B. Mark Welch, L. May & E. J. Walsh / Evolving rotifers, evolving science
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Montero-Pau, J., Serra, M. & Gómez, A. Diapausing egg banks, lake size, and genetic diversity in the rotifer Brachionus plicatilis Müller (Rotifera, Monogononta). Hydrobiologia 796, 77–91 (2017). https://doi.org/10.1007/s10750-016-2833-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-016-2833-4