Abstract
This study provides the first analysis of genome size diversity in Monogonont rotifers. Measurements were made using flow cytometry, with Drosophila melanogaster and chicken erythrocytes as internal standards. Nuclear DNA content (“2C”—assuming diploid genomes) in eight different species of four different genera ranged almost fourfold, from 0.12 to 0.46 pg. A comparison with previously published values for Bdelloid rotifers suggested that the genomes of Monogononts are significantly smaller than those of Bdelloids. When compared to other Metazoans, Monogonont rotifers seem to have relatively small genomes. For instance, the C-values of the two species with the smallest genomes, Brachionus dimidiatus and Synchaeta pectinata, were only 0.06 and 0.085 pg, respectively. Various explanations for genome size diversity within Monogononta are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Monogonont rotifers are the most species-rich order within the Phylum Rotifera, comprising about 1,500 morphologically distinct species (Segers, 2007). This figure does not include an even higher number of cryptic species, which may be nested within each morphospecies (e.g., Gomez et al., 2002; Gilbert & Walsh, 2005; Suatoni et al., 2006). Monogononts are also extremely diverse in their ecology, inhabiting various aquatic and semi-aquatic habitats (fresh and saline water), tolerating a wide range of abiotic conditions, and exhibiting an enormous repertoire of feeding modes, from suspension feeding to carnivory, even cannibalism, and highly specialized predator–prey adaptations. Genetic and broad-scale phylogenetic characterization of Monogonont rotifers is still in its infancy, even though some groups are reasonably well studied (e.g., the Brachionus plicatilis species complex). By contrast, almost nothing is known about key genomic parameters, such as genome size. To date, genome sizes have been estimated for nine different Bdelloid species (Pagani et al., 1993; Welch & Meselson, 1998, 2003) and only one Monogonont rotifer (Welch & Meselson, 1998).
Estimates of genome size (i.e., “2C values,” in the case of diploid species/tissues, or the C-value, if referring to haploid genome size) are interesting for various theoretical and practical reasons. First, they can serve as decision basis for planned genome sequencing projects in a particular taxonomic group. Species with small genome sizes are often given priority in such projects, for economic reasons. Second, genome size can be viewed as a complex organismal trait, which is shaped by various selective forces. For instance, genome size can be influenced by developmental constraints, such that it tends to be reduced in species with fast development (e.g., Gregory & Johnston, 2008). On the other hand, relaxed selection, e.g., due to low effective population sizes, may facilitate a proliferation of selfish genetic elements and, thus, lead to increases in genome size (Lynch & Conery, 2003). Third, genome size measurements can provide information on recent polyploidization events or “cryptopolyploidization,” e.g., if closely related species exhibit discontinuous genome size variation that is/is not reflected in the karyotype (e.g., Gregory & Hebert, 2002). Finally, the so-called C-value paradox, the enormous diversity in nuclear genome size in animals and plants, a long-standing puzzle to biologists, is still not completely understood (Gregory, 2001). The full extent of genome size variation among organisms may even not have received full appreciation, since most genome size databases tend to have a strong bias toward a few intensely studied taxa (e.g., vertebrates), while genome size diversity in other groups, such as most invertebrates, has been largely ignored.
There are mainly two established types of methods for genome size determination. The first group comprises methods in which samples are prepared by squashing tissues on microscopic slides, followed by fixation and staining of nuclei and densitometric examination of staining intensity. Such methods make use of classical DNA stains, such as the Feulgen reaction (Pagani et al., 1993; Hardie et al., 2002), or fluorescent dyes (e.g., Welch & Meselson, 2003). Another widely used method is flow cytometry. Here tissues or whole animals are homogenized in a special buffer, stained with fluorescent dyes and analyzed in a flow cytometer. The basic principle of flow cytometry implies that cell homogenate is conveyed in a very narrow fluid stream, and fluorescence signals, corresponding to individual nuclei, are counted and measured in their intensity. In all methods, Feulgen densitometry, its fluorescent analogs, and in flow cytometry internal standards (using tissues of organisms with known genome size) are usually processed at the same time, which allows an estimate of the absolute genome size and makes comparisons of results of different studies possible.
This study provides the first assessment of genome size diversity in Monogonont rotifers, covering eight species of four different genera. Measurements were made using flow cytometry. In addition, several basic life history variables were measured (body size, egg size, and relative egg size) and tested for a potential relationship with genome size. In rotifers, these variables are loosely correlated with development time, a life history variable, which is suspected to constrain genome size in other animals (e.g., Gregory & Johnston, 2008).
Materials and methods
Clonal cultures of rotifers were established from the offspring of single amictic females. In total, 20 different rotifer clones were analyzed (usually two clones per species or strain). The studied rotifers comprised eight morphologically recognizable species: Brachionus calyciflorus, B. plicatilis, B. angularis, B. quadridentatus, B. dimidiatus, Synchaeta pectinata, Keratella cochlearis (a spineless form), and Asplanchna priodonta. The Florida strain of B. calyciflorus was obtained from J.J. Gilbert (Dartmouth College, NH, USA) and has been characterized in Gilbert & Walsh (2005). The Egelsee strain of B. calyciflorus was isolated from Lake Egelsee (Austria) and was also used in previous studies (e.g., Stelzer, 2008). The Russian strain of B. plicatilis has long been used in various laboratory studies (e.g., Snell & Stelzer, 2005) and was originally isolated from the Azov Sea (Russia). All other rotifer clones were isolated in spring 2009 from various small ponds in the vicinity of the Institute for Limnology, Mondsee (Austria), except for the Bogoria strain of B. plicatilis and B. dimidiatus, which were hatched from a sediment sample of Lake Bogoria (East Africa).
To obtain large amounts of biomass, rotifers were cultured in flasks of 1 l volume. The Brachionus species were cultured in glass bottles, aerated with sterile air through a glass tube. All other rotifers (Asplanchna, Keratella, and Synchaeta) were cultured in Fernbach flasks, which were not aerated, but allowed good gas exchange due to a large surface-to-volume ratio. Rotifers were fed with the algae Tetraselmis suecica (B. plicatilis and B. dimidiatus), Chlamydomonas reinhardii (B. calyciflorus, B. angularis, and B. quadridentatus), and Cryptomonas erosa (S. pectinata and K. cochlearis), and the carnivorous rotifer Asplanchna priodonta was fed with a small, obligately asexual B. calyciflorus strain (body length: ~140 μm). When cultures reached population densities of 10–100 individuals per ml, rotifer biomass was harvested with 30-μm sieves (or 200 μm, for Asplanchna), resuspended in sterile culture medium, and starved for 2 h. This procedure was repeated twice to insure that rotifers completely emptied their guts. Cleaned rotifer biomass was resuspended in 10 ml ice-cold Galbraith buffer (DeSalle et al., 2005), concentrated by centrifugation (1 min at 1,000×g) and buffer was removed to 0.3–1 ml (depending on the amount of initial biomass). This procedure usually resulted in a harvest of 10–20 μl pellet (concentrated biomass), suspended in 0.3–1 ml Galbraith buffer.
Concentrated biomass was ground on ice with 100 strokes in a 1-ml Dounce tissue homogenizer, to free individual nuclei. Large debris was removed by filtration through a 35-μm mesh nylon sieve. A detergent-trypsin method was used to prepare the nuclei for flow cytometric analysis (Mullen, 2004). Briefly, 100 μl of the homogenized cell suspension was digested by addition of 450 μl of 0.003% Trypsin for 10 min at room temperature. To prevent further degradation, 0.05% trypsin inhibitor was added (this solution also included 0.01% RNAse A) and the samples were incubated for another 10 min. Finally, samples were stained with propidium iodide at a concentration of 50 μg/ml. Stained samples were kept on ice in the dark and analyzed after 1–3 h. Flow cytometric analysis was performed in a FacsCalibur flow cytometer (BD Biosciences) using the manufacturer’s instructions. Each sample was analyzed until a pre-specified number of 10,000 counted events (i.e., particles registered by the fluorescence detectors) were reached. In most runs, Drosophila melanogaster (strain ISO-1) was used as an internal standard for genome size. The heads of ten female Drosophila were homogenized in 0.5 ml Galbraith buffer with 15 strokes in the Dounce tissue homogenizer, and 100 μl of this homogenate was Trypsin-digested and stained as described above. Rotifer samples and Drosophila standards were first run separately, to identify the position of the diploid peaks, and then combined into the same sample and measured again. If there was overlap between fluorescence peaks of rotifers and Drosophila (due to similar genome size), chicken erythrocytes (from the DNA Quality control Kit of BD Biosciences) were used as an alternative internal standard. The genome of chicken is about seven times larger than that of Drosophila melanogaster (Gregory, 2009).
Body size and egg size were measured using inverted microscopy at 100-fold magnification (Asplanchna) and 200-fold magnification (all other rotifers). The Brachionus species and Keratella were fixed in 4% formalin before the measurements, whereas Asplanchna and Synchaeta were measured alive in a plankton compression chamber according to Uhlig (Hydrobios, Kiel, Germany), since both species showed strong distortion of their body shape upon treatment with formalin. Measurements on body volume were done according to the recommendations by Ruttner-Kolisko (1977), except for Keratella cochlearis and Brachionus angularis, which were measured according to Walz (1983). Relative egg size was calculated as the ratio between egg volume and body volume (Walz, 1995).
Results
Nuclear genome sizes (“2C values”) in the eight different rotifer species ranged almost fourfold, from 0.12 to 0.464 pg (Table 1). Interestingly this range was encompassed within the genus Brachionus, with B. dimidiatus having the smallest and B. plicatilis (Bogoria strain) having the largest genome (Table 1; Fig. 1a, b). Genome sizes of the other genera, Asplanchna, Synchaeta, and Keratella, fell within this range. Replicate measurements, either from different extractions of the same clone or from different clones of the same species/population, gave highly consistent results, as can be seen by the small standard deviations (Table 1). Further support for the accuracy of the flow cytometric measurements was provided by tests in which the two reference species, Drosophila and chicken, were combined in the same sample. These measurements gave two clear peaks, with the fluorescence intensity of chicken erythrocytes being seven times larger than that of Drosophila (Fig. 1b), a value consistent with the expected ratio of 6.94 (C-values of Chicken 1.25 pg and Drosophila 0.18 pg; according to Gregory 2009).
Examples of genome size measurements using flow cytometry. The intensity of the fluorescence signal is shown on the x-axis. The counts on the y-axis refer to the fluorescence signals emitted by individual nuclei. a The two smallest genomes of this study: Brachionus dimidiatius and Synchaeta pectinata, with Drosophila as internal standard. b The largest genome of this study: Brachionus plicatilis (Bogoria strain), with Drosophila and chicken erythrocytes as internal standards. c Haploid male peak and diploid female peak of a culture of B. plicatilis (Russian strain). d Haploid male peak and diploid female peak of a culture of B. calyciflorus (Florida strain)
Closer inspection of the fluorescence histograms of some species revealed a second, usually smaller peak in addition to the diploid peak (Fig. 1c, d), which had almost exactly half of the fluorescence intensity. These additional peaks were most likely caused by haploid males, which were present in large quantities in some of the cultures. Such “male peaks” were observed in cultures of B. plicatilis, B. calyciflorus, and Asplanchna. Another interesting observation was that the genome size of the Bogoria strain of Brachionus plicatilis was almost exactly twice the size of the Russian strain (Table 1).
In order to reveal potential constraints on genome size, I checked for correlations of three basic life history variables with genome size. None of these correlations were significant: body size (Spearman’s rank correlation, ρ = −0.018, df = 8, P = 0.9602), egg size (ρ = 0.05, df = 7, P = 0.8984), relative egg size (ρ = 0.15, df = 7, P = 0.7001). However, there was a significant correlation between relative egg size and body size (ρ = −0.867, df = 7, P = 0.0025), with small rotifers having the largest relative egg sizes. The latter result is in agreement with other studies in Monogonont rotifers (Walz, 1995; Stelzer, 2005).
Discussion
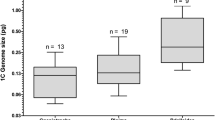
This study provides the first analysis of genome size diversity in Monogonont rotifers. To date, genome sizes have only been measured in six Bdelloid rotifer species (Pagani et al., 1993; Welch & Meselson, 1998, 2003) and only one Monogonont rotifer (a member of the Brachionus plicatilis species complex; Welch & Meselson, 1998). As all previous studies measured absolute genome sizes and involved internal standards with reference organisms of known genome size, an inter-study comparison is possible. This comparison indicates that the genome sizes of Monogononts are on average smaller than those of Bdelloid rotifers (Fig. 2). The median nuclear DNA content of Monogononts was 0.294 pg, while that of Bdelloids was 1.5 pg (compiled from Pagani et al., 1993 and Welch & Meselson, 1998, 2003). This difference is statistically significant (Mann–Whitney Rank Sum Test, T = 143, n = 9.11, P < 0.001). It should be noted, however, that there is overlap between both groups, as the largest Monogonont genome (Brachionus plicatilis, 0.7 pg, in Welch & Meselson, 1998) was larger than the smallest Bdelloid genome (Adineta sp., 0.5 pg, in Welch & Meselson, 2003). To get a more complete picture, it will be necessary in future studies to measure many more species/genera of both Bdelloid and Monogonont rotifers.
Nuclear DNA content of Monogonont and Bdelloid rotifers. All values for Monogononts were estimated in this study (see Table 1), except one value for B. plicatilis, which was taken from Welch & Meselson (1998). All genome sizes of Bdelloid rotifers were compiled from previous studies (Pagani et al., 1993; Welch & Meselson, 1998, 2003)
A comparison of Monogonont rotifers with representatives of other animal phyla suggests that monogonont genomes range at the lower end of Metazoan genome sizes. The C-values of the two smallest genomes in this study were 0.06 and 0.085 pg, for Brachionus dimidiatus and Synchaeta pectinata, respectively (C-values calculated from nuclear DNA content assuming diploid tissues). Examples for other invertebrates with similarly small genomes are: Paraturbanella teissieri (Gastrotricha, 0.05 pg), Trichoplax adhaerens (Placozoa, 0.04 pg), Dysidae crawshagi (Sponge, 0.06), and several plant-parasitic nematodes, some of which have C-values as low as 0.02 pg (all values taken from the animal genome size database; Gregory, 2009).
The small genome sizes in Monogonont rotifers could be a result of selection for short egg development times (i.e., high rates of cell divisions during embryonic development). Analogous constraints have been suggested in Drosophila, where species with small body size and fast juvenile development tend to have smaller genomes (Gregory & Johnston, 2008). Monogonont rotifers are among the fastest growing Metazoans, in terms of the intrinsic rate of population increase (Bennett & Boraas, 1989), and rapid juvenile and embryonic development are major components of this fast growth. In addition, cell divisions occur only during embryonic development. In this study of Monogonont rotifers, I found no evidence for a relationship between body size, egg size, and relative egg size on genome size. However, it may nevertheless be worthwhile to directly measure embryonic development times and to test for a relationship with genome size, since these life history variables are only loosely correlated with embryonic development times (Walz, 1995; Stelzer, 2005).
An unexpected pattern was detected in the two Brachionus plicatilis strains: The nuclear DNA content of the Bogoria strain was almost exactly twice of that of the Russian strain. It is tempting to speculate that this is the result of whole-genome duplication. The only previous measurement of genome size in a Monogonont rotifer was done on another member of the B. plicatilis species complex (Welch & Meselson, 1998) and was 0.7 pg. This is roughly three times higher than the value of the Russian strain. Thus, in future studies it might be very interesting to more rigorously examine genome size diversity within the Brachionus plicatilis species complex.
References
Bennett, W. N. & M. E. Boraas, 1989. A demographic profile of the fastest growing metazoan: a strain of Brachionus calyciflorus (Rotifera). Oikos 55: 365–369.
DeSalle, R., T. R. Gregory & J. S. Johnston, 2005. Preparation of samples for comparative studies of arthropod chromosomes: visualization, in situ hybridization, and genome size estimation. Methods in Enzymology 395: 460–488.
Gilbert, J. J. & E. J. Walsh, 2005. Brachionus calyciflorus is a species complex: mating behavior and genetic differentiation among four geographically isolated strains. Hydrobiologia 546: 257–265.
Gomez, A., M. Serra, G. R. Carvalho & D. H. Lunt, 2002. Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera). Evolution 56: 1431–1444.
Gregory, T. R., 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Reviews 76: 65–101.
Gregory, T. R., 2009. Animal Genome Size Database. http://www.genomesize.com.
Gregory, T. R. & P. D. N. Hebert, 2002. Genome size estimates for some oligochaete annelids. Canadian Journal of Zoology 80: 1485–1489.
Gregory, T. R. & J. S. Johnston, 2008. Genome size diversity in the family Drosophilidae. Heredity 101: 228–238.
Hardie, D. C., T. R. Gregory & P. D. N. Hebert, 2002. From pixels to picograms: a beginners’ guide to genome quantification by Feulgen image analysis densitometry. Journal of Histochemistry and Cytochemistry 50: 735–749.
Lynch, M. & J. S. Conery, 2003. The origins of genome complexity. Science 302: 1401–1404.
Mullen, P., 2004. Flow cytometric DNA analysis of human cancer cell lines. In Langon, S. P. (ed.), Methods in Molecular Medicine, Vol. 88: Cancer Cell Culture: Methods and Protocols. Humana Press Inc, Totowa, NJ: 248–255.
Pagani, M., C. Ricci & C. A. Redi, 1993. Oogenesis in Macrotrachela quadricornifera (Rotifera, Bdelloidea) I. Germarium eutely, karyotype and DNA content. Hydrobiologia 255(256): 225–230.
Ruttner-Kolisko, A., 1977. Suggestions for biomass calculations of plankton rotifers. Archiv fuer Hydrobiologie Beihefte 8: 71–76.
Segers, H., 2007. Annotated checklist of the rotifers (Phylum Rotifera), with notes on nomenclature, taxonomy and distribution. Zootaxa 1564: 1–104.
Snell, T. W. & C. P. Stelzer, 2005. Removal of surface glycoproteins and transfer among Brachionus species. Hydrobiologia 546: 267–274.
Stelzer, C. P., 2005. Evolution of rotifer life histories. Hydrobiologia 546: 335–346.
Stelzer, C. P., 2008. Obligate asex in a rotifer and the role of sexual signals. Journal of Evolutionary Biology 21(1): 287–293.
Suatoni, E., S. Vicario, S. Rice, T. W. Snell & A. Caccone, 2006. An analysis of species boundaries and biogeographic patterns in a cryptic species complex: the rotifer Brachionus plicatilis. Molecular Phylogenetic and Evolution 4: 86–98.
Walz, N., 1983. Continuous culture of the pelagic rotifers Keratella cochlearis and Brachionus angularis. Archiv für Hydrobiologie 98: 70–92.
Walz, N., 1995. Rotifer populations in plankton communities: energetics and life history strategies. Experientia 51: 437–453.
Welch, D. M. & M. Meselson, 1998. Measurements of the genome size of the Monogonont rotifer Brachionus plicatilis and the Bdelloid rotifers Philodina roseola and Habrotrocha constricta. Hydrobiologia 387(388): 395–402.
Welch, D. M. & M. Meselson, 2003. Oocyte nuclear DNA content and GC proportion in rotifers of the anciently asexual class Bdelloidea. Biological Journal of the Linnean Society 79: 85–91.
Acknowledgments
I would like to thank Johanna Schmidt and Anneliese Wiedlroither for technical assistance in cultivation of rotifers and morphological measurements, Peter Stadler for introducing me to the use of the flow cytometer, Christian Schlötterer for supplying a culture of Drosophila melanogaster, and Steve Omondi for supplying sediment from Lake Bogoria. Financial support was provided by FWF grant P20735-B17.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Guest editors: N. Walz, R. Adrian, J.J. Gilbert, M.T. Monaghan, G. Weithoff & H. Zimmermann-Timm / Rotifera XII: New aspects in rotifer evolution, genetics, reproduction, ecology and biogeography
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Stelzer, CP. A first assessment of genome size diversity in Monogonont rotifers. Hydrobiologia 662, 77–82 (2011). https://doi.org/10.1007/s10750-010-0487-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10750-010-0487-1