Abstract
Apoptosis and clearance of dead cells is highly evolutionarily conserved from nematode to humans, which is crucial to the growth and development of multicellular organism. Fail to remove apoptotic cells often lead to homeostasis imbalance, fatal autoimmune diseases, and neurodegenerative diseases. Small ubiquitin-related modifiers (SUMOs) modification is a post-translational modification of ubiquitin proteins mediated by the sentrin-specific proteases (SENPs) family. SUMO modification is widely involved in many cellular biological process, and abnormal SUMO modification is also closely related to many major human diseases. Recent researches have revealed that SUMO modification event occurs during apoptosis and clearance of apoptotic cells, and plays an important role in the regulation of apoptotic signaling pathways. This review summarizes some recent progress in the revelation of regulatory mechanisms of these pathways and provides some potential researching hotpots of the SUMO modification regulation to apoptosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis is an evolutionarily conserved cell progress from nematode to mammals, which is of great importance for organisms normal development, failure to apoptotic cells clearance can result in the release of toxins and inflammation of the surrounding environment (Elliott and Ravichandran 2010; Munoz et al. 2005). Caenorhabditis elegans is an important genetic system for studying the phagocytic mechanism of apoptotic cells, and the molecular mechanism of apoptotic cell clearance has been extensively studied in C. elegans (Reddien and Horvitz 2004; Lettre and Hengartner 2006). Two major signaling pathways trigger apoptotic cell death: the mitochondrial (internal) pathway and the death receptor (external) pathway. Normally, apoptotic cells are recognized and degraded by phagocytes (such as macrophages) before releasing potentially toxic substances (Galluzzi et al. 2012). Defective apoptotic cell clearance can lead to a variety of diseases, including autoimmune diseases, neurodegenerative disorders.
Small ubiquitin-related modifier (SUMO) is involved in diverse cellular processes and disorder of sumoylation is implicated in the pathogenesis of human diseases. Proteins are the physical basis of life and the basic unit of the function and structure of organisms. The Small Ubiquitin-like Modifier (SUMO) post-translational modify a large number of proteins that function in diverse biological processes, including transcription, chromatin remodeling, DNA repair, and mitochondrial dynamics. SUMOylation is also a dynamic process, which can be reversed by a family of conserved Sentrin/SUMO-specific proteases (SENPs). Post-translational modification of ubiquitin-like proteins has been shown to be involved in many regulatory processes of cellular activities, such as cell cycle, signaling, immune recognition, apoptosis, cell proliferation and differentiation, inflammation, and DNA damage repair (Gareau and Lima 2010; Zhao et al. 2004; Muller et al. 2001).
In this review, we briefly describe the process of cell death and the SUMOylation then discuss the relationship between apoptosis and ubiquitination-like modification.
Mechanisms of apoptosis
Lockshin and Williams originally defined programmed cell death in the context of insect development. Subsequently, Kerr et al. noticed by ultrastructural analysis two morphologically different types of cell death in humans, apoptosis, and necrosis (Lockshin and Williams 1965; Kerr 1971).
During apoptosis, the cells shrink with integral but ruffling plasma membranes, and nuclei are condensed and fragmented (Saraste and Pulkki 2000). Apoptosis is one of the important ways of programmed cell death, the process can be divided into several important steps, including the initiation and transmission of death signals, the triggering of death procedures, and the clearance of apoptotic cells (Elmore 2007). Apoptosis is mediated by a series of caspases that function in the cascades, and caspase 3 or caspase 7 is responsible for killing cells at the end of caspase 3 or caspase 7. Apoptosis involves not only the mechanism of killing cells, but also the release of “find me” signals to recruit macrophages and the exposure of “eat me” signals to bind macrophages, which efficiently promotes the elimination of apoptotic cells (deCathelineau and Henson 2003; Medina and Ravichandran 2016).
Apoptotic cells are cleared by specialized phagocytes, such as macrophages and dendritic cells, or by non-specialized phagocytes, such as epithelial cells. This process plays a very important role in organ formation and tissue dynamic balance (Arandjelovic and Ravichandran 2015). Apoptotic cells are swiftly phagocytosed by macrophages to prevent the release of intracellular components, timely and effective elimination of apoptotic cells can also prevent the body from producing inflammation or autoimmune diseases. In contrast, failure to remove the apoptotic cell often leads to a variety of severe chronic inflammatory or fatal autoimmune diseases (systemic lupus erythematosus), as well as neurodegenerative diseases (Alzheimer's disease) (Wang and Yang 2016; Nagata et al. 2010).
Apoptosis can be defined as cell death accompanied by activation of the caspase protease. Apoptosis is strictly controlled by multiple factors, stages, and genes, which is regulated by the following signal transduction pathways: mitochondrial pathway, death receptor signaling pathway, and so on.
Apoptosis involves the activation of caspases, which can destroy all morphological changes of cell death. Caspases are cysteine proteases that are specific to aspartic acid residues in the substrate. Although there are at least 17 different caspases in mammals, we focus on only a small number of them, and the activation of these caspases is at least partially understood, and the role of these caspases in cell death has been determined.
The mitochondrial pathway of apoptosis
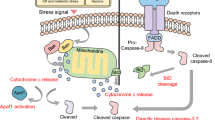
When cells are regulated by internal apoptotic stimulators (Oncogene activation, DNA damage, hypoxia, growth factor damage, endoplasmic reticulum stress, etc.), the mitochondrial pathway, an intrinsic pathway of cell apoptosis, is activated and thus cell apoptosis is triggered. This process increases mitochondrial membrane permeability and releases Cyt-C into the cytoplasm to interact with Apaf1, leading to a series of Caspase cascade reactions.
The mitochondrial pathway of apoptosis, also known as the intracellular pathways of apoptosis, is triggered by internal apoptotic stimulators (Oncogene activation, DNA damage, hypoxia, growth factor damage, endoplasmic reticulum stress, etc.) (Bratton and Salvesen 2010; Bratton et al. 2001). During apoptosis, cytochrome c is released from the mitochondria into the cytoplasm and binds to apoptotic protease activator 1 (APAF1, CED-4 in C.elegans), which triggers the hydrolysis of the Apaf1 cofactor dATP to dADP (Adrain et al. 1999). The subsequent exchange of dADP and exogenous dATP resulted in the aggregation of 7 apaf1-datp-cyto-chromeon-c cells into an active apoptotic body (Kim et al. 2006). At the apoptotic corpuscle center, APAF1 activates the inactive caspase 9 monomer (Thornberry and Lazebnik 1998). In healthy cells, cytochrome c is present only in the mitochondrial membrane gap. In addition to cytochrome c, two other pro-apoptotic proteins are released during this process: Smac (also known as diablo) and Omi(also known as HtrA2) (Thornberry and Lazebnik 1998; Zou et al. 1999). MOMP is an event strictly controlled by members of the Bcl2 family. There are three main types of Bcl2 proteins: pro-apoptotic effectors ( Bax and Bak); Anti-apoptotic Bcl2 proteins (such as Bcl2, Bcl-xl, Mcl1); And bh3-specific proteins (activating pro-apoptotic effector factor and neutralizing anti-apoptotic Bcl2 protein). CED-9 protein in C.elegans is similar to human anti-apoptotic protein Bcl2/ Bcl-xl. And EGL-1 is similar to several bh3-specific proteins. In the embryo of C. elegans, CED-9 and CED-4 are located in the mitochondria, and CED-4 is isolated in the mitochondrial membrane. When cell apoptosis occurs, egl-1 is activated by transcription and interacts with CED-9 to convert CED-4 from the mitochondrial surface to the peri-nuclear region, thus promoting the activation of CED-3 caspase of cells and eventually leading to cell death (Tait and Green 2010; Tait et al. 2010; Yuan and Horvitz 1990; Yan et al. 2005).
The CED-3 caspase, a homologous gene of human CASP2 (caspase 2), is involved in the positive regulation of apoptosis. Studies have shown that Dicer/dcr-1 ribonuclease is a substrate of CED-3. In the process of apoptosis, CED-3 can convert DCR-1 into deoxyribonuclease and initiate chromosome breakage. Moreover, mitochondrial fission proteins drp-1/Drp1 and multi-transmembrane protein CED-8/Xk1 also undergo CED-3 cleavage, and the cleaved DRP-1 and CED-8 promote the exposure of phosphatidylserine (PtdSer) as a "eat me" signal on the surface of apoptotic cells (Nakagawa, et al. 2010; Chen et al. 2013a; Breckenridge et al. 2008).
Death receptor signaling pathway
The death receptor (DRs) pathway is one of the important pathways mediating apoptosis and can trigger apoptosis. DRs belongs to tumor necrosis factor receptor (TNFR) and has an extracellular domain rich in Cys and an intracellular death domain (DD). When the death receptor binds to a specific death ligand, the extracellular death signal activates the intracellular apoptosis mechanism through DRs, inducing apoptosis. The known death receptors-ligands are Fas (APO-1, CD95)-FasL (CD95L), TNFR1-TNF, TRAILR1/2-TRAIL. (I) In the process of Fas-FasL signal transduction, FADD, Daxx, FILP, and other related proteins are recruited by intracellular terminal DD; (II) after TNFR1-TNF binding, THE TNFR1 DD is induced to aggregate and recruit the bridging protein TRADD, which can recruit signal molecules such as TRAF2, RIP and FADD; (III) TRAILR1/2-TRAIL binds to FADD in cancer cells via DD. FADD then recruits pro-caspase8 through the death effect domain DED to form the death-induced signaling complex (DISC), and the pro-caspase8 on the DISC cuts itself into the active caspase8, thus mediating apoptosis (Carrington et al. 2006).
In type I cells, activated caspase-8 promotes apoptosis by dividing and activating caspase-3 and caspase-7. In type II cells, XIAP inhibits active caspases, thereby preventing apoptosis. Caspase-8 mediated cleavage activates Bid, which in turn activates Bax and Bak to promote MOMP. IAP antagonists Smac and Omi were subsequently reused to neutralize XIAP and allow apoptosis to continue. The signaling pathway downstream of TNFR1 is more complex and may lead to three different outcomes: survival, apoptosis, or necrosis. TNFR1-associated death domain protein (TRADD) does not directly activate caspase-8, but it can serve as a membrane-binding scaffold for additional signaling molecules, including kinases, receptor interaction protein 1 (RIP1), and ubiquitin-ligases TRAF2 and cIAP1/2. When the ubiquitination of RIP1 is inhibited by the ubiquitin ligase contained in complex I (such as cIAP1/2) or directly deubiquitinated by enzymes such as CYLD, TNFR1 ligation can lead to cell death. RIP1 and TRADD are released from TNFR1 to form a series of dynamic cytosolic signaling platforms (collectively, complex II) that may lead to cell apoptosis and necrosis. Cytoplasmic TRADD recruits FADD (via dd-dd interaction) and activates caspase-8. In this structure, the formation of complex II leads to apoptosis and death (Newton et al. 2012; Jost et al. 2009).
Apoptotic cell recognition
The most-studied ‘eat-me signal’ is phosphatidylserine (PS), a phospholipid that is exposed on the surface of dying cells (Leventis and Grinstein 2010). In normal cells, PS is mainly located in the inner leaflet of the cell membrane, and it is continuously transferred from the outer lobule to the inner lobule of the cell membrane by amino-phosphatidyltransferase TAT-1, to maintain its asymmetric distribution on the cell membrane (Fadok et al. 1992; Williamson and Schlegel 2003). Not only PS and calreticulin, an intracellular chaperone protein involved in the transport of correctly folded proteins through the endoplasmic reticulum, may become exposed on the surface of apoptotic cells (Darland-Ransom et al. 2008; Kuraishi et al. 2007). In C.elegans, TTR-52 is an extracellular protein similar to trans-thymosin secreted by non-apoptotic intestinal cells, which can recognize and bind to the exposed PS on the surface of apoptotic cells. TTR-52 promotes the movement of PS vesicles from apoptotic cells to phagocytes together with the ABC transporter protein, CED-7 (Mapes et al. 2012). Meanwhile, TTR-52 interacts with the extracellular domain of phagocytic receptor CED-1, consequently delivers the “eat me” signal to phagocytes, accomplishing the clearance of apoptotic cells (Zhou et al. 2001). The CED-1 homolog in Drosophila, Draper (Drpr) has also been shown to play a role in the elimination of apoptotic cells by hemocytes/macrophage both in vitro and in vivo (Manaka et al. 2004). In addition to the phagocytic receptor CED-1, the phosphatidylserine receptor psr-1, an evolution-conserved PtdSer receptor, also mediates cell corpse recognition by PS, and then promotes the elimination of apoptotic cells through CED-2, CED-5, and CED-12 signaling pathways (Wang et al. 2003).
Cell corpse clearance
The clearance of apoptotic cells involves several dynamic steps including: mutual recognition between apoptotic cells and the phagocytic receptors, cytoskeleton rearrangement in phagocytes, phagosome formation after endocytosis, and the ultimate degradation of apoptotic cells in phagocytes (Li et al. 2013). Apoptotic cell clearance is highly evolutionarily conserved, but this process is extremely redundant and complex in mammalian systems, with multiple molecules playing a similar role in apoptotic cell clearance (Wang et al. 2003). By studying the genetics of programmed cell death in model animal, the molecular mechanism of apoptotic cells clearance has been preliminarily understood.
Phagocytic cytoskeletal rearrangement and phagosome sealing
On the one hand, CED-1 interacts with CED-6 to recruit clathrin CHC-1 and the multi-subunit adaptor protein AP2, forming a complex to mediate actin cytoskeleton rearrangement (Chen et al. 2013b); on the other hand, AP2 also interact with LST-4 and DYN-1 to initiate the cell intima into the "phagocytic cup", subsequently causes cytoskeletal rearrangement and promotes the extension of phagocytic pseudopodia (Yu et al. 2006). Another pathway that controls the phagocytosis of apoptotic cells by adjacent cells is composed of CED-2, CED-5, CED-10, CED-12, and PSR-1 (Hsu and Wu 2010). PSR-1 is involved in the recognition and binding of PtdSer to initiate apoptosis cell clearance. Non-receptor tyrosine kinase SRC-1 can combine with INA-1 and CED-2 to transmit phagocytosis signal to the downstream. CED-2 interacts with CED-5, and CED-5 can form a complex with CED-12 to activate CED-10 as a guanosine exchange factor. CED-10 is a homologous of human RacI guanosylates hydrolase, which can promote the rearrangement of the cytoskeleton and the phagocytosis of apoptotic cells (Liu and Hengartner 1999).
Under the action of these two pathways, phagocytes of C. elegans continuously extended to completely envelop apoptotic cells in the membrane of phagocytic cells, and finally form phagocytes. Phagocytic pseudopodia fusion completely encloses the phagocytic body of apoptotic cells. PtdIns(4,5)P2 and PtdIns3P phosphoinositol play a very important role in the closure of phagocytes. PtdIns(4,5)P2 is catalyzed by PtdIns4P 5 kinase ppk-1 and is mainly concentrated in the unsealed phagocytes. PtdIns3P phosphatase MTM-1 (myotubularin) and PIKI-1 of type II phosphoinositol 3 kinase synergistically controlled the content of PtdIns3P in unsealed phagocytes. Then, the SNX9 family protein LST-4 was recruited to the phagocytosis by detecting PtdIns(4,5), P2, PtdIns3P, MTM-1, and LST-4 further recruited DYN-1 to complete phagocytosis (Cheng et al. 2015).
Phagosome formation and maturation
Phagocytic maturation includes the processes from the generation of phagocytes to the fusion of lysosomes and phagocytes (Vieira et al. 2002). This process starts immediately after the phagocytic sealing, even before this. After the phagosome is sealing, it falls off from the cell membrane, and fuses with the early endosome, late endosome, and lysosome successively, and fuses with the lysosome to form phagolysosome. At this time, various hydrolytic enzymes in the lysosome form an acidic environment to degrade the phagosome. It has been found that Rab GTPases (Rab1/2/3/4/5/7/11/14) play a very important role in the fusion of phagocytes and lysosomes and the acidification of phagolysosomes, such as regulating the clearance of apoptotic cells and phagocytosis (Stuart et al. 2007; Garin et al. 2001). It has been shown that early phagocytosis recruited RAB-5 protein in mammals, Drosophila, and C. elegans, while RAB-7 participates in the late stage of phagosome maturation and mediates phagocytic and lysosomal fusion (Vieira et al. 2002; Kinchen and Ravichandran 2008; Kinchen et al. 2008).
RAB-5 binds to early phagocytes, which may promote the production of PtdIns3P by activating type III phosphoinositol 3 kinase VPS-34 (Kinchen et al. 2008). Effector molecules of PtdIns3P, such as SNX-1 and SNX-6, are recru ited into the phagolysosome and mediate the recycling of the phagocytic receptor CED-1 (Norris et al. 2017). Then, with the help of SAND-1, CCZ-1, and GTPase activating protein (GAP) TBC-2, the phagocytes undergo a gradual maturation phase that exchanges the combination from RAB-5 to RAB-7 through the Rab exchange mechanism (Li et al. 2009; Kinchen and Ravichandran 2010). The other two RAB GTPases, RAB-14 and UNC-108(the RAB-2 homolog of C.elegans), and the activated molecule gopher 1 of UNC-108 play a role in the maturation of phagocytes earlier than RAB-7. Rab-14 and UNC-108 are likely to facilitate the recruitment of lysosomes to phagocytes, while RAB-7 acts as an anchor to fuse lysosomes and phagocytes. In addition, the HOPS complex and the small GTPase ARL-8 are also essential for the formation of phagocytic lysosomes. Finally, the lysosomal lysine/arginine transporter LAAT-1 maintains the lysosome formation, the lysosomal cathepsin L(CPL-1), and type II DNA enzyme (NUC-1) mediate the digestion and degradation of apoptotic cells (Liu et al. 2012; Xu et al. 2014).
Synopsis of SUMO conjugation and deconjugation
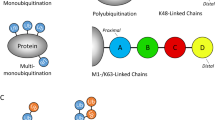
SUMOylation, similar to ubiquitination, can covalently modify lysine residues of target proteins and is an important way of reversible post-translational modification of target proteins. It has been found that SUMOylation has biological significance for the function, localization, and stabilization of target proteins, thus affecting cell growth, differentiation, and apoptosis. Single/poly SUMOylation involves many biological processes, such as regulation of gene transcription, protein–protein interactions, nucleoplasmic transport, signal transduction, and protein degradation. In addition to the involvement of sumoylation in the regulation of protein–protein and protein-DNA interactions, the most important thing is that more studies have found that SUMOylation of proteins may be related to the occurrence and development of human diseases, such as cancer, neurodegenerative diseases, and immune diseases (Flotho and Melchior 2013; Nie and Boddy 2016).
SUMO proteins
SUMO is an 11-kDa protein that is a highly conserved family and widely found in eukaryotes. Yeast, nematode, and fruit fly have only one SUMO gene, while the human genome encodes four different SUMO proteins: SUMO1-SUMO4 (Table 1). SUMO1-SUMO3 are widely expressed, while SUMO4 is only expressed in specific tissues, such as kidney and spleen (Johnson 2004). Moreover, the mature SUMO2 and SUMO3 share 97% sequence identity to form poly-SUMO chains, while their homology with SUMO1 is only 50% (Johnson 2004). SUMO1 is very different from SUMO2/3 in function, and previous data have shown that they have distinct and overlapping sets of target proteins and the substrate protein it binds to in the body is also different (Saitoh and Hinchey 2000; Tatham et al. 2001; Vertegaal et al. 2006).
The same substrate protein can be modified by different subtypes of SUMO protein, and even modified by the same subtype of SUMO protein can lead to different biological effects due to the various stimulus factors. SUMO family is conjugated to proteins which regulate a variety of cellular processes such as nuclear transport, transcription, chromosome segregation, and DNA repair, suggesting that SUMO signaling plays important roles in diverse processes by altering proteins activities, their ability to interact with other proteins, or their subcellular localization (Dohmen 2004).
SUMO chains
In the ubiquitin system, different chain connections produce a variety of signals that determine the fate of the modified protein. The SUMOylation includes activation, binding, connection, modification, and dissociation. All SUMO proteins present as immature precursors, requiring the maturation of SUMO proteases to expose the C-terminal diglycine (GG) motif.
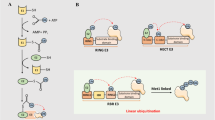
SUMOylation affects the activation, interaction, subcellular localization, and stability of substrate proteins. In recent years, a series of new ubiquitin ligases (E3), known as ULS(E3-S) or STUbL, have been discovered that recognize SUMO proteins and associate SUMO modification with the ubiquitin/protease system. SUMO's connection to the substrate also requires a series of enzymatic cascades, including E1 activase (Sae1/Sae2 heterodimer in humans), E2 conjugating enzyme (Ubc9 in humans), and E3 ligase (Guo et al. 2007). Connections between SUMO and other SUMO lysine residues (K) contribute to the formation of substrate connections to the SUMO chain (poly-SUMOylation) (Pfander et al. 2005). SUMO modification is highly dynamic and reversible (Mendler et al. 2016). The SUMO monomer or SUMO chains can be unbound to the protein by the SUMO isopeptidase (ULP, SENP, SUSP). There are three different SUMO proteins in mammals, but only one SUMO homolog in C. elegans, named SUMO-1. The homologs of E1 activase in C. elegans are AOS-1/UBA-2, E2 conjugating enzyme homologs is UBC-9, and E3 ligase homologs are GEI-17 and MMS-21 respectively.
Like ubiquitin, mature SUMO molecules require activation to modify the substrate, a process performed by the SUMO activating enzyme. SUMO activating enzyme (E1) is a heterodimer formed by AOS1 (SAE1, Sua1) and UBA2 (SAE2), whose N-terminal and C-terminal are similar to the corresponding structures of ubiquitin-activating enzyme (Dohmen et al. 1995). SUMO conjugating enzyme (E2) through transesterification reaction, the activated SUMO molecules are transferred from the SUMO activating enzyme UBA2 subunit to the SUMO conjugating enzyme (E2), also known as UBC9, to form the SUMO-UBC9 sulfur ester intermediate. As one of the main nuclear proteins, UBC9 can locate both the cytoplasmic and cytoplasmic sides of the nuclear pore complex (NPC) (Bossis and Melchior 2006). Several proteins are currently identified as SUMO E3 ligases, and a comprehensive biochemical and structural analysis of three types of proteins (sp-ring (Siz/Pias) family, RanBP2, and ZNF451 family) was performed to provide insight into their SUMO catalytic patterns (Pichler et al. 2002) (Table 1).
SUMO proteases
The SENP family mediates the de-SUMOylation modification of substrate proteins. Immature SUMO protein has an extended chain of 2–11 amino acids at its C-terminal, and the extended chain needs to be cut off by the SUMO specific protease SENP (sentrin-specific protease), exposing diglycine (Gly-Gly) to become stable form, so as to conduct subsequent modification of the target protein. The human genome encodes six SENPs: SENP1, SENP2, SENP3, SENP5, SENP6, and SENP7(Yeh et al. 2000). Their C-terminal is a conserved protease catalytic domain with about 200 amino acids. However, the N-terminal of different SENPs proteins varies greatly from sequence to structure, which may be related to their ability to precisely act on different target proteins (Garvin et al. 2013; Hang and Dasso 2002; Nishida et al. 2001). In C. elegans, the SENP family consists of four SUMO proteases (ubiquitin-like proteases, ULPs) ULP-1, ULP-2, ULP-4, and ULP-5. Ulp-2 has been reported to inhibit E-cadherin and promotes its recruitment to adhesive connections. Moreover, ULP-4 has been reported to deSUMOylate HMGS-1 to control mevalonate pathway activity during aging (Tsur et al. 2015; Sapir et al. 2014).
SENP1 exists in the nuclear pore and nuclear membrane, which can travel between the cytoplasm and the nucleus. SENP2 is the nuclear membrane related protease, which mainly present in the nucleus, but experiments have shown that it is also present in the cytoplasm (Itahana et al. 2006). SENP3 and SENP5, which may be likely to modify SUMO2/3. SENP6 and SENP7 are mainly located in the nucleus (Garvin et al. 2013), which are significantly different from SENP1, SENP2, SENP3, and SENP5 in structure, have an additional ring structure in their catalytic region, mainly in the nuclear material, and de-SUMOylation of SUMO2/3 (Nayak and Muller 2014).
One substrate protein provides more free SUMO protein to modify other substrates as well as de-SUMOylation. The restrictive SUMO precursor, the mature SUMO subtype, and the SENP family determine the SUMO modification balance (Hickey et al. 2012).
The crosstalk between SUMO modification and apoptosis
SUMO modification is a post-translational modification that affects protein stability, transcriptional activity, protein–protein interactions, and intracellular localization of target proteins. It has been reported that SUMOylation can regulate the activity of NR4A1. NR4A is a nuclear receptor protein family, whose members act as sensors in the cellular environment to regulate metabolism, proliferation, migration, apoptosis, autophagy, and other processes, thus inducing autophagy-dependent cell death. The process is outlined in Fig. 1. Moreover, mutation of three ubiquitin-like sites of NR4A1 can increase its transcriptional activity, alter its intracellular distribution, and more importantly, its ability to induce autophagy death is impaired (Zárraga-Granados et al. 2020; Zhang et al. 2017).
Myeloid cell leukemia 1(MCL1), an anti-apoptotic protein that belongs to the BCL-2 family, often keeps a high expression in many cancers during oncogenesis. The stability of MCL1 is maintained by SUMOylation at its K234 and K238 sites, which inhibits Tripartite motif-containing 11(TRIM11) -mediate ubiquitination of MCL1 and apoptosis of cancer cells (Li et al. 2020).
DAXX(death domain-associated protein) was a kind of FAS binding protein and could act as a regulator of JNK-mediated apoptosis (Baltimore et al. 2000). DAXX has been shown to mediate apoptosis through exogenous death receptor pathways and to regulate gene expression by interacting with multiple DNA-binding transcription factors (TFs) and chromatin related proteins as transcriptional co-inhibitors or co-activators (Hollenbach et al. 2002; Li et al. 2000). In addition, it was reported that DAXX contains SUMO-interacting motifs (SIMs) (Santiago et al. 2009). The Phosphorylation of the Daxx-SIM promotes SUMOylation by binding SUMO-1 to SUMO-2/3, thereby enhances stress-induced cell apoptosis by repressing the antiapoptotic genes (Chang et al. 2011). Some studies have reported that in the case of cell damage caused by reactive oxygen species (ROS), the SUMO ligase PIAS1 can activate the JNK signaling pathway and inhibit the expression of anti-apoptotic genes (Feligioni et al. 2011).
Many recent studies have reported the use of SUMO modification for the treatment of diseases, especially cancer. As the only E2 coupling enzyme in the ubiquitin-like modified pathway, UBC-9 has been widely used in many clinical treatments. Studies have shown that 2-D08, as a small molecular reagent to prevent the process from UBC9 to substrates, may induce the apoptosis of AML cells mediated by ROS accumulation through the de-SUMOylation of NOX2 (Kim et al. 2014; Zhou et al. 2019). In addition, p53, as a tumor suppressor protein, is associated with post-translational modifications of various proteins. When p53 is transferred to the nucleus and modified by SUMO, the transcriptional expression of pro-apoptotic gene Bax can be up-regulated (Wasiak et al. 2007). In the rat model of intervertebral disc degeneration (IDD), the positive expression of SUMO2/3 protein was higher. In addition, silencing the SUMO2 gene reduces the expression level and phosphorylation level of p53, while increasing the expression level of CDK2/4 and CyclinB1. Therefore, SUMO2 influences the apoptosis and senescence of nucleus pulposus cells in the IDD rat model by mediating the p53 signaling pathway (Jin et al. 2018).
SUMO modification disorders are likely to result in human diseases. A large number of studies have shown that the SUMO modification is in an unbalanced condition during the development of various cancers (Rabellino et al. 2017). Besides, accelerated apoptosis is involved in neurodegenerative diseases, such as Alzheimer's disease and Parkinson's disease. In addition, SUMO modification protects the body from viruses. However, viral proteins could compete with host cells for the SUMOs to infect bodies. From this perspective, SUMO modification alters virus-host interactions (Zhu et al. 2019; Isaacson and Ploegh 2009).
Conclusion and prospect
Over the past few decades, we've learned how apoptosis is triggered, what molecules are activated to kill cells, and how phagocytes recognize apoptotic cells. Apoptosis plays an important role in the development of organisms and maintains homeostasis. Timely and effective elimination of apoptotic cells can also prevent the body from producing inflammation or autoimmune diseases due to the release of their harmful contents. Thus, deregulating signaling pathways that trigger cell death can lead to cancer and autoimmune diseases (too little cell death) and degenerative diseases (too much cell death). Certain regulatory pathways can activate or inhibit cell death, depending on the developmental environment. The specific activation of cell death during development need to be further studied, thus help us to understand how cell death is precisely controlled in the treatment of human diseases. By studying the intersection of developmental pathways with conserved core apoptosis mechanisms, we can learn how cancer cells develop resistance to cell death, or how to prevent cell death in neurodegenerative diseases such as Parkinson's disease.
The way a cell dies can have important effects on neighboring cells, and sometimes even the entire organism. The signals that cause death may vary with different types of cell death. The death pathways are clearly interrelated. For example, autophagic death is usually enhanced by caspase activation, which is antagonistic in caspase-dependent necrosis. The interconnections between these pathways may provide several possible mechanisms for cell death procedures (Abraham et al. 2004; Nicotera and Melino 2004).
Over the past 20 years, our understanding of the SUMO mechanism of proteins has expanded greatly. SUMO molecules participate in post-translational modification of proteins, but do not mediate proteasome degradation of target proteins. Instead, they reversibly modify target proteins and participate in their positioning and functional regulation. However, it is not clear how SUMO uses its limited number of enzymes to achieve substrate specificity, compared to a large number of substrates. Moreover, the regulation mechanism of SUMO needs to be further studied.
At present, some progress has been mentioned in the role of SUMOylation in apoptosis. SUMOylation plays an important role in the functional modification of protein substrates. Therefore, elucidating the role of SUMOylation in apoptosis is of great significance for some chronic inflammatory diseases caused by apoptosis and helps us to understand the pathogenesis. However, the current studies on the role of SUMOylation in apoptosis mainly focus on the process of the occurrence of apoptosis, while little is known about the SUMOylation involved in the process of cell corpse clearance. Therefore, the study of SUMOylation in the process of apoptotic cell clearance is helpful for us to further reveal the pathogenesis of chronic inflammatory response caused by apoptotic clearance disorder and provide a new theoretical basis for the development of drugs in clinical practice.
References
Abraham MC, Shaham SJTICB (2004) Death without caspases, caspases without death. Trends Cell Biol 14(4):184–193
Adrain C, Slee EA, Harte MT et al (1999) Regulation of apoptotic protease activating factor-1 oligomerization and apoptosis by the WD-40 repeat region. J Biol Chem 274(30):20855–20860
Arandjelovic S, Ravichandran KS (2015) Phagocytosis of apoptotic cells in homeostasis. Nat Immunol 16(9):907–917
Baltimore D, Chang HY, Khosravi-Far R et al (2000) Daxx, a Fas-binding protein that activates JNK and apoptosis, US. Cell 89(7):1067–1076
Bossis G, Melchior F (2006) Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell 21(3):349–357
Bratton SB, Salvesen GS (2010) Regulation of the Apaf-1-caspase-9 apoptosome. J Cell Sci 123(Pt 19):3209–3214
Bratton SB, Walker G, Srinivasula SM et al (2001) Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J 20(5):998–1009
Breckenridge DG, Kang BH, Kokel D et al (2008) Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell 31(4):586–597
Carrington PE, Sandu C, Wei YF et al (2006) The structure of FADD and its mode of interaction with procaspase-8. Mol Cell 22(5):599–610
Chang CC, Naik MT, Huang YS et al (2011) Structural and functional roles of Daxx SIM phosphorylation in SUMO paralog-selective binding and apoptosis modulation. Mol Cell 42(1):62–74
Chen YZ, Mapes J, Lee ES et al (2013) Caspase-mediated activation of Caenorhabditis elegans? CED-8 promotes apoptosis and phosphatidylserine externalization. Nat Commun 4:2726
Chen D, Jian Y, Liu X et al (2013) Clathrin and AP2 are required for phagocytic receptor-mediated apoptotic cell clearance in Caenorhabditis elegans. PLoS Genet 9(5):e1003517
Cheng S, Wang K, Zou W et al (2015) PtdIns(4,5)P(2) and PtdIns3P coordinate to regulate phagosomal sealing for apoptotic cell clearance. J Cell Biol 210(3):485–502
Darland-Ransom M, Wang X, Sun CL et al (2008) Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320(5875):528–531
deCathelineau AM, Henson PM (2003) The final step in programmed cell death: phagocytes carry apoptotic cells to the grave. Essays Biochem 39:105–117
Dohmen RJ (2004) SUMO protein modification. Biochim Biophys Acta 1695(1–3):113–131
Dohmen RJ, Stappen R, McGrath JP et al (1995) An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J Biol Chem 270(30):18099–18109
Elliott MR, Ravichandran KS (2010) Clearance of apoptotic cells: implications in health and disease. J Cell Biol 189(7):1059–1070
Elmore S (2007) Apoptosis: a review of programmed cell death. Toxicol Pathol 35(4):495–516
Fadok V, Voelker DR, Campbell PA (1992) Exposure of phosphatidylserine on the surface of poptotic lymphocytes triggers specific recognition and removal by macrophages. J Immunol 148:22–29
Feligioni M, Brambilla E, Camassa A et al (2011) Crosstalk between JNK and SUMO signaling pathways: deSUMOylation is protective against H2O2-induced cell injury. PLoS ONE 6(12):e28185
Flotho A, Melchior F (2013) Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385
Galluzzi L, Vitale I, Abrams JM et al (2012) Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ 19(1):107–120
Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol 11(12):861–871
Garin J, Diez R, Kieffer S et al (2001) The phagosome proteome: insight into phagosome functions. J Cell Biol 152(1):165–180
Garvin AJ, Densham RM, Blair-Reid SA et al (2013) The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep 14(11):975–983
Guo B, Yang SH, Witty J et al (2007) Signalling pathways and the regulation of SUMO modification. Biochem Soc Trans 35(Pt 6):1414–1418
Hang J, Dasso M (2002) Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem 277(22):19961–19966
Hickey CM, Wilson NR, Hochstrasser M (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13(12):755–766
Hollenbach AD, Mcpherson CJ, Mientjes EJ et al (2002) Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein. Dek 115(Pt 16):3319
Hsu TY, Wu YC (2010) Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr Biol 20(6):477–486
Isaacson MK, Ploegh HL (2009) Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5(6):559–570
Itahana Y, Yeh ET, Zhang Y (2006) Nucleocytoplasmic shuttling modulates activity and ubiquitination-dependent turnover of SUMO-specific protease 2. Mol Cell Biol 26(12):4675–4689
Jin LZ, Lu JS, Gao JW (2018) Silencing SUMO2 promotes protection against degradation and apoptosis of nucleus pulposus cells through p53 signaling pathway in intervertebral disc degeneration. Biosci Rep 38(3):20171523
Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73:355–382
Jost PJ, Grabow S, Gray D et al (2009) XIAP discriminates between type I and type II FAS-induced apoptosis. Nature 460(7258):1035–1039
Kerr JF (1971) Shrinkage necrosis: a distinct mode of cellular death. J Pathol 105(1):13–20
Kim H-E, Du F, Fang M et al (2006) Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci USA 102:17545–17550
Kim YS, Keyser SG, Schneekloth Jr. JS (2014) Synthesis of 2’,3’,4’-trihydroxyflavone (2–D08), an inhibitor of protein sumoylation. Bioorg Med Chem Lett 24(4):1094–1097
Kinchen JM, Ravichandran KSJNRMCB (2008) Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol 9(10):781–795
Kinchen JM, Ravichandran KS (2010) Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells. Nature 464(7289):778-U157
Kinchen JM, Doukoumetzidis K, Almendinger J et al (2008) A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 10(5):556–566
Kuraishi T, Manaka J, Kono M et al (2007) Identification of calreticulin as a marker for phagocytosis of apoptotic cells in Drosophila. Exp Cell Res 313(3):500–510
Lettre G, Hengartner MO (2006) Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol 7(2):97–108
Leventis PA, Grinstein S (2010) The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys 39:407–427
Li H, Leo C, Zhu J et al (2000) Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol Cell Biol 20(5):1784–1796
Li W, Zou W, Zhao D et al (2009) C elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development 136(14):2445–2455
Li Z, Lu N, He X et al (2013) Monitoring the clearance of apoptotic and necrotic cells in the nematode Caenorhabditis elegans. Methods Mol Biol 1004:183–202
Li S, Wang J, Hu G et al (2020) SUMOylation of MCL1 protein enhances its stability by regulating the ubiquitin-proteasome pathway. Cell Signal 73:109686
Liu QA, Hengartner MO (1999) The molecular mechanism of programmed cell death in C. elegans. Ann N Y Acad Sci 887:92–104
Liu B, Du H, Rutkowski R et al (2012) LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science 337(6092):351–354
Lockshin RA, Williams CM (1965) Programmed cell death–I. Cytology of degeneration in the intersegmental muscles of the pernyi silkmoth. J Insect Physiol 11:123–133
Manaka J, Kuraishi T, Shiratsuchi A et al (2004) draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by drosophila hemocytes/macrophages. J Biol Chem 279(46):48466–48476
Mapes J, Chen YZ, Kim A et al (2012) CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr Biol 22(14):1267–1275
Medina CB, Ravichandran KS (2016) Do not let death dous part: ‘find-me’ signals in communication between dying cells and the phagocytes. Cell Death Differ 23(6):979–989
Mendler L, Braun T, Muller S (2016) The ubiquitin-like SUMO system and heart function from development to disease. Circ Res 118(1):132–144
Muller S, Hoege C, Pyrowolakis G et al (2001) Sumo, ubiquitin’s mysterious cousin. Nat Rev Mol Cell Biol 2(3):202–210
Munoz LE, Herrmann M, Gaipl US (2005) An impaired detection and clearance of dying cells can lead to the development of chronic autoimmunity. Z Rheumatol 64(6):370–376
Nagata S, Hanayama R, Kawane K (2010) Autoimmunity and the clearance of dead cells. Cell 140(5):619–630
Nakagawa A, Shi Y et al (2010) Caspase-dependent conversion of dicer ribonuclease into a death-promoting deoxyribonuclease. Science 328(5976):327–334
Nayak A, Muller S (2014) SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15(7):422
Newton K, Dixit VM (2012) Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol 4(3):a006049–a006049
Nicotera P, Melino G (2004) Regulation of the apoptosis-necrosis switch. Oncogene 23(16):2757–2765
Nie M, Boddy MN (2016) Cooperativity of the SUMO and ubiquitin pathways in genome stability. Biomolecules 6(1):14
Nishida T, Tanaka H, Yasuda HJEJB (2001) A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem 267(21):6423–6427
Norris A, Tammineni P, Wang S et al (2017) SNX-1 and RME-8 oppose the assembly of HGRS-1/ESCRT-0 degradative microdomains on endosomes. Proc Natl Acad Sci USA 114(3):E307–E316
Pfander B, Moldovan G, Sacher M et al (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436(7049):428–433
Pichler A, Gast A, Seeler JS et al (2002) The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108(1):1–120
Rabellino A, Andreani C, Scaglioni PP (2017) The role of PIAS SUMO E3-ligases in cancer. Cancer Res 77(7):1542–1547
Reddien PW, Horvitz HR (2004) The engulfment process of programmed cell death in caenorhabditis elegans. Annu Rev Cell Dev Biol 20:193–221
Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275(9):6252–6258
Santiago A, Godsey AC, Hossain J et al (2009) Identification of two independent SUMO-interacting motifs in Daxx: Evolutionary conservation from Drosophila to humans and their biochemical functions. Cell Cycle 8(1):76–87
Sapir A, Tsur A, Koorman T et al (2014) Controlled sumoylation of the mevalonate pathway enzyme HMGS-1 regulates metabolism during aging. Proc Natl Acad Sci USA 111(37):E3880–E3889
Saraste A, Pulkki K (2000) Morphologic and biochemical hallmarks of apoptosis. Cardiovasc Res 45(3):528–537
Stuart LM, Boulais J, Charriere GM et al (2007) A systems biology analysis of the Drosophila phagosome. Nature 445(7123):95–101
Tait SWG, Green DR (2010) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11(9):621–632
Tait SWG, Parsons MJ, Llambi F et al (2010) Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell 18(5):1–813
Tatham MH, Jaffray E, Vaughan OA et al (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276(38):35368–35374
Thornberry NA, Lazebnik Y (1998) Caspases: enemies within. Science 281(5381):1312–1316
Tsur A, Bening A-S, Broday L (2015) ULP-2 SUMO protease regulates E-cadherin recruitment to adherens junctions. Dev Cell 35(1):63–77
Vertegaal AC, Andersen JS, Ogg SC et al (2006) Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics 5(12):2298–2310
Vieira OV, Botelho RJ, Grinstein S (2002) Phagosome maturation: aging gracefully. Biochem J 366(Pt 3):689–704
Wang X, Yang C (2016) Programmed cell death and clearance of cell corpses in Caenorhabditis elegans. Cell Mol Life Sci 73(11–12):2221–2236
Wang X, Wu YC, Fadok VA et al (2003) Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science 302(5650):1563–1566
Wasiak S, Zunino R, Biology HMMJJOC (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 177(3):439–450
Williamson P, Schlegel RAJBEBA (2003) Transbilayer phospholipid movement and the clearance of apoptotic cells 1585(2–3):53–63
Xu M, Liu Y, Zhao L et al (2014) The lysosomal cathepsin protease CPL-1 plays a leading role in phagosomal degradation of apoptotic cells in Caenorhabditis elegans. Mol Biol Cell 25(13):2071–2083
Yan N, Chai JJ, Lee ES et al (2005) Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature 437(7060):831–837
Yeh ET, Gong L, Kamitani T (2000) Ubiquitin-like proteins: new wines in new bottles. Gene 248(1–2):1–14
Yu X, Odera S, Chuang CH et al (2006) C elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell 10(6):743–757
Yuan JY, Horvitz HR (1990) The Caenorhabditis elegans genes ced-3 and ced-4 act cell autonomously to cause programmed cell death. Dev Biol 138(1):33–41
Zárraga-Granados G, Muciño-Hernández G, Sánchez-Carbente M et al (2020) The nuclear receptor NR4A1 is regulated by SUMO modification to induce autophagic cell death. PLoS ONE 15:e0222072
Zhang L, Xie F, Zhang J et al (2017) SUMO-triggered ubiquitination of NR4A1 controls macrophage cell death. Cell Death Differ 24(9):1530–1539
Zhao Y, Kwon SW, Anselmo A et al (2004) Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J Biol Chem 279(20):20999–21002
Zhou Z, Hartwieg E, Horvitz HRJC (2001) CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104(1):43
Zhou P, Chen X, Li M et al (2019) 2–D08 as a SUMOylation inhibitor induced ROS accumulation mediates apoptosis of acute myeloid leukemia cells possibly through the deSUMOylation of NOX2. Biochem Biophys Res Commun 513(4):1063–1069
Zhu Z, Chu H, Wen L et al (2019) Targeting SUMO modification of the non-structural protein 5 of Zika virus as a host-targeting antiviral strategy. Int J Mol Sci 20(2):392
Zou H, Li Y, Liu X et al (1999) An APAF1 Cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem 274(17):11549–11556
Acknowledgements
This work was partially supported by the National Natural Science Foundation of China (Grant No. 31671439 to H. Xiao), Natural Science Foundation of Shaanxi Province, China (Grant No. 2020JM-271 to H. Xiao), National Natural Science Foundation of China Youth Program (Grant No. 31801164 to Q. Zheng).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, P., Jing, H., Wang, Y. et al. SUMO modification in apoptosis. J Mol Histol 52, 1–10 (2021). https://doi.org/10.1007/s10735-020-09924-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10735-020-09924-2