Abstract
We investigated the effect of overexpressing Fragaria vesca L. cv. Rügen S-adenosyl-l-methionine synthase (FvSAMS) and decarboxylase (FvSAMDC) genes on control and salt stressed Nicotiana benthamiana Domin plants. According to previous studies the overproduction of both proteins enhances the abiotic stress tolerance of plants, but the two enzymes have not yet been studied in one experimental system. We found that the transgenic plants subjected to long-term salt stress displayed higher levels of tolerance than the wild type (WT). In contrast to several earlier studies no antagonistic effect between ethylene and polyamine biosynthesis was observed in our experimental system. Overexpression of FvSAMDC had higher impact on the plant physiological parameters both in control and salt stress conditions, than that of FvSAMS. Based on the data measured in the FvSAMDC lines there appears to be a positive correlation between the free polyamine levels and the proline content as well as the amount of ethylene, while there is a negative correlation between the free polyamine levels and the lignin content in the plants exposed to salt stress. The transformation vectors contained the CaMV35S promoter, the coding sequence of FvSAMS and FvSAMDC fused with synthetic green fluorescent protein (sGFP). We detected the subcellular localization of both enzymes and examined the possible stress induced changes in their distribution. In the case of FvSAMS::sGFP nuclear, nucleolar, cytoplasmic (near to the plasmalemma), plastid membrane, whereas in FvSAMDC::sGFP nuclear and homogenous cytoplasmic localization was detected. Therefore, SAM is assumed to be produced in situ for numerous biochemical reactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Different abiotic stresses lead to severe economic losses each year (Mahajan and Tuteja 2005). High salt levels cause ion toxicity (mainly Na+), hyperosmotic stress, and secondary stresses (i. e. oxidative damage) (Zhu 2002; Chinnusamy et al. 2006). Both ethylene and polyamines (putrescine, spermidine and spermine) play an important regulatory role in the biotic and abiotic stress responses of plants (Müller and Munné-Bosch 2015; Romero et al. 2018) and in larger quantities they have a positive effect on the salt tolerance of plants (Jang et al. 2012; Peng et al. 2014; Shen et al. 2014). The antioxidant enzyme activity is growing on the effect of adequate concentration of both ethylene and polyamine, by this way they increase the homeostasis of reactive oxygen species (ROS) and regulate the balance of Na/K, enhancing the adaptation of plants to the salinity environment (Saha et al. 2015; Tao et al. 2015). S-adenosyl-l-methionine (SAM) is a common precursor of polyamine and ethylene biosynthesis. The biosynthesis of ethylene proceeds through the following steps: SAM → 1-aminocyclopropane-1-carboxylic acid (ACC) → ethylene catalyzed by ACC synthase (ACS) and ACC oxidase (ACO) enzymes. At the same time the SAM decarboxylase (SAMDC) converts SAM to decarboxylated SAM, which acts as an aminopropyl group donor in the synthesis of polyamines. Numerous earlier studies reported on the antagonistic effect of ethylene and polyamine metabolism in response to various biotic and abiotic stresses (Li et al. 2004; Nambeesan et al. 2012; Yu et al. 2016). Therefore, we also set out to study how the ‘SAM flux’ and ‘SAM deficiency’ influenced the ratio of ethylene and polyamine in response to salt stress. S-adenosyl-l-methionine synthase produces S-adenosyl-l-methionine from l-methionine and ATP. SAM is the second most frequently used enzyme substrate after ATP (Cantoni 1975) and can serve as a methyl group source for amino, ribosyl and aminopropyl groups (Fontecave et al. 2004). Furthermore, SAM is needed for the methylation of DNA, RNA, proteins, lipids, lignin, pectin, volatiles, flavonoids, nicotianamine and biotin (Roeder et al. 2009; Gong et al. 2014). Since DNA methylation causes genes to be shut down, SAM is definitely considered to be a gene expression regulator. According to estimates 95% of SAM is used for methylation and only 3–5% is decarboxylated (Merali and Clarkson 2004). SAM is not only the precursor of ethylene and polyamines, but the secondary metabolite lignin, too. Lignin is one of the main metabolic consumers of SAM. In mto3 Arabidopsis thaliana genotype a point mutation of the SAMS3 gene, causing a single amino acid change in the ATP binding domain, is responsible for the mto3 phenotype. There is no modification either in SAMS3 expression pattern or in the amount of SAMS protein produced but there is a decrease in SAMS enzyme activity and in the total amount of SAM and lignin content compared to the wild type (WT), providing evidence for the impact of SAMS3 on lignin biosynthesis (Shen et al. 2002). Several former studies reported an increase in the amount of SAMS mRNA in response to salt stress in rose periwinkle, wheat and ginseng (Schröder et al. 1997; Pulla et al. 2009; Kamal et al. 2012). The overexpression of SAMS positively influenced the abiotic stress tolerance of tomato, tobacco and A. thaliana (Qi et al. 2010; Gong et al. 2014; Guo et al. 2014; Kim et al. 2015; Ma et al. 2017).
SAMDC forms a small gene family in A. thaliana—the 4 members of which show high similarity to each other (Franceschetti et al. 2001)—and is a strictly regulated enzyme (Hu et al. 2005). The in vivo production of the enzyme can be both transcriptionally and translationally regulated with the latter being directly influenced by polyamines (Pegg 1986; Kusano and Suzuki 2015). SAMDC participates in developmental processes, cell differentiation and it is synthetized as a proenzyme, which undergoes an autocatalytic cleavage. SAMDC functions as a dimer in mammals containing an alfa and a beta chain. In numerous higher organisms, its autocatalysis and decarboxylation are stimulated by putrescine, but in plants SAMDC is highly active in the absence of putrescine as well (Bennett et al. 2002). SAM decarboxylase plays a pivotal role in regulating spermidine and spermine biosynthesis in numerous plant species, since the decarboxylated SAM (dcSAM) produced by the SAMDC acts as a substrate for both spermidine and spermine synthase (Kusano et al. 2008). Embryonic lethality was observed in the double mutant of bud2 and samdc1, underlying the fundamental role of SAMDC in plant embryogenesis (Ge et al. 2006). The up- and down-regulation of the SAMDC showed that the ethylene and polyamine biosynthesis has antagonistic impact on each other in A. thaliana (Hu et al. 2006). Furthermore, the overexpression of carnation SAMDC resulted in elevated salt stress tolerance in transgenic tobacco plants (Wi et al. 2006) and the NaCl stress increased the amount of MdSAMDC1 and AtSAMDC1 mRNA in apple and A. thaliana (Urano et al. 2003; Liu et al. 2008). The silencing of SAMDC increased salt sensitivity in tobacco, at the same time dcSAM deficiency had a positive effect on the biomass production and stem length of transgenic plants (Mellidou et al. 2016).
Our goal was to elucidate the functions of these two enzymes (FvSAMS and FvSAMDC) both under control and salinity stress conditions, since they have not been investigated simultaneously in one experimental system. Earlier studies investigated the effect of overexpressing SAMS or SAMDC during abiotic stresses in Nicotiana tabacum L., Solanum lycopersicum L. and A. thaliana (Boerjan et al. 1994; Roy and Wu 2002; Waie and Rajam 2003; Wi et al. 2006, 2014; Cheng et al. 2009; Peremarti et al. 2009; Qi et al. 2010; Gong et al. 2014; Kim et al. 2015; Ma et al. 2017). Studying concomitant activity of the two enzymes in the same experimental system allows the plant physiological parameters investigated to be related to one another quantitatively, making it possible to build a clearer picture with regard to the role of the two enzymes in salt stress response. In addition, we planned to study the subcellular localization of FvSAMS::sGFP, FvSAMDC::sGFP and the possible changes in response to 10 mM NaCl stress. The SAMS is considered an enzyme with cytosolic localization in plants (Schröder et al. 1997; Ravanel et al. 1998; Hanson and Roje 2001), whereas it was found to show nucleocytoplasmic distribution in mammals (Reytor et al. 2009). Furthermore, in earlier studies SAMDC activity was found in the chloroplasts, mitochondria and cytosol of plants (Yamanoha and Cohen 1985; Torrigiani et al. 1986), while only cytoplasmic localization was detected in animals (Gritli-Linde et al. 1995).
Material and methods
Plant material, vector constructs and bioinformatic analysis
For the identification of sequences, the NCBI database was used. The main ORF of FvSAMS (XP_004288342.1) codes for 394 amino acids and shows 95% similarity with AtSAMS1 (NP_171751.1), 93% with AtSAMS2 (NP_192094.1) and 90% with AtSAMS3 (NP_181225.1). The main ORF of FvSAMDC (XP_011462957.1) corresponds to 360 amino acids and its similarity is 64% to AtSAMDC1 (NP_001154585.1), 66% to AtSAMDC2 (NP_197099.1), 68% to AtSAMDC3 (NP_001189972.1) and 43% to AtSAMDC4 (NP_197394.1). The leaf tissue of Fragaria vesca L. cv. Rügen was used for the isolation of RNA and cDNA was synthesized by reverse transcription according to the method described below. The cDNA sequences of FvSAMS and FvSAMDC were amplified with the primer pairs designed for the main ORFs and the CaMV35S promoter was amplified with specific primer pairs (Table S2). NetNES 1.1 (La Cour et al. 2004), TargetP 1.1 (Emanuelsson et al. 2007), cNLS Mapper (Kosugi et al. 2009), GPS-SNO 1.0 (Xue et al. 2010), Nucleolar localization sequence Detector (NoD) (Scott et al. 2011) and PHOSIDA (Gnad et al. 2011) applications were used for bioinformatic analysis of the sequences. The cDNA fragments were ligated into pGWB405 (AB294429.1) and the CaMV35S was ligated into pGWB604 (AB543113.1) binary vector according to manufacturer’s protocol (Invitrogen™ Gateway®, ThermoFisher Scientific, Waltham, USA). The pGWB405 contained a constitutive CaMV35S promoter and the synthetic green fluorescent protein (sGFP) reporter gene while the pGWB604 carried only the sGFP. The FvSPDS::sGFP construct used for microscopic analysis was built by us using the pGWB405 vector. The insert codes for the spermidine synthase gene isolated from F. vesca L. cv. Rügen. FvSPDS::sGFP fusion protein showed only cytoplasmic localization and was used as a cytoplasmic control. The FvSAMS-22 and FvSAMS-25 as well as the FvSAMDC-73 and FvSAMDC-76 transgenic lines are referred to hereinafter as FvSAMS and FvSAMDC lines.
DNA, RNA isolation and cDNA synthesis
The isolation of genomic DNA, plasmid DNA and total RNA was carried out by the CTAB method, PureYield™ Plasmid Miniprep System Kit (Promega, Madison, USA) and the Total RNA Mini Kit (Plant) (Geneaid®, New Taipei City, Taiwan), respectively, according to the manufacturer’s protocols. cDNA was synthetized from total RNA by using Oligo (dT)18 primer according to the protocol of the Revert-Aid First Strand cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, USA), which was used later for the qPCR analysis.
Agrobacterium mediated plant transformation and transient expression assay
The Agrobacterium tumefaciens GV3101 strain was used for transformation of the Nicotiana benthamiana plants based on the method described by Clemente (2006). The A. tumefaciens GV3101 strain contained the pGWB405::FvSAMS, the pGWB405::FvSAMDC and the pGWB604::CaMV35S vectors. The same constructions were used for the in vivo infiltration of the 35-day-old N. benthamiana plants based on the method described by Li (2011).
Plant growth conditions
The seeds of two transgenic T1 lines from each construct (FvSAMS-22, FvSAMS-25 and FvSAMDC-73, FvSAMDC-76) were germinated on MS medium supplemented with 75 µg/ml kanamycin while the wild type N. benthamiana seeds on MS medium without antibiotics, respectively. Fourteen days after gemination (14 DAG), 32 seedlings were germinated of each transgenic line and wild type were either transferred onto half strength MS medium without antibiotics or onto the same medium supplemented with 0.58 g/l (10 mM) NaCl. The plants and the explants were kept in 16 h light and 8 h dark periods at 23 °C. Samples were collected 85 days after sowing. We investigated the effect of 17.54 g/l (300 mM) NaCl on the survival of N. benthamiana leaf discs on same plant growth conditions than in the case of 10 mM NaCl stress. All plant physiology parameters were measured from N. benthamiana plant samples grown in control and 10 mM NaCl stress conditions. All photographs were taken with Fuji FinePix S6500fd camera.
Chlorophyll a, b and relative chlorophyll content measurements
Chlorophyll a and b as well as the total chlorophyll contents were measured according to the method published by Porra et al. (1989). The chlorophyll a (Ca) and chlorophyll b (Cb), as well as the total chlorophyll (Ct = Ca + Cb) contents were determined based on the absorbance measured at 645 nm (Abs.645) and at 663 nm (Abs.663) by using NanoDrop ND-1000 UV/Vis spectrophotometer (NanoDrop Technologies, USA) and calculated according to the formulas: Ca = 0.0127 (Abs.663) − 0.00269 (Abs.645); Cb = 0.0229 (Abs.645) − 0.00468 (Abs.663). ImageJ 1.51j8 software was used for determining the relative chlorophyll content of leaf discs (Schneider et al. 2012). Leaves of wild type N. benthamiana were used for calibration between 0 and 100% chlorophyll content (chlorophyll-free and control chlorophyll samples). We calculated the relative chlorophyll content from the measured values of 12 leaf discs.
Histochemical detection of H2O2 and O2−
The histochemical visualization of H2O2 and O2− was carried out according to the method described by Kumar et al. (2014). The chlorophyll-free leaves were fixed in 60% (v/v) glycerol.
Determination of H2O2 content
The H2O2 content was determined according to Velikova et al. (2000). The absorbance of the samples was measured at 390 nm by using the WPA Biotech Photometer 1101 (Cambridge, UK). For plotting the standard curve dilutions of 30% (w/w) H2O2 (Sigma-Aldrich, Saint Louis, USA) were used.
Electrolyte leakage measurement
The electrolyte leakage measurement was carried out according to the method described by Rizhsky et al. (2002). The conductivity of samples was measured by using Radelkis Conductometer Type OK-104 (Budapest, Hungary).
Determination of proline content
The semi-quantitative determination of proline content was performed by the isatin paper assay of Ábrahám et al. (2010) protocol. l-proline (Sigma-Aldrich, Saint Louis, USA) solutions were used for plotting the standard curve. Photographs were evaluated using ImageJ 1.51j8 software (Schneider et al. 2012).
Lignin measurement
The lignin content of the shoots was determined by the acetyl bromide method described by Moreira-Vilar et al. (2014). The absorbance of the samples was measured at 280 nm with WPA Biotech Photometer 1101 (Cambridge, UK). The standard curve was plotted using alkali lignin (Sigma-Aldrich, Saint Louis, USA).
Ethylene analysis
Ethylene release of the leaf discs was measured according to the method described by Langebartels et al. (1991). We put five leaf discs of 9 mm diameter on filter paper soaked in 1 ml 50 mM MES/NaOH (pH 5.6) and 2% (w/v) sucrose solution. We placed the scrolled filter papers into glass tubes covered by silica septum then incubated at 25 °C for 1 h. 1 ml of gas was injected into the Shimadzu GC-14A gas chromatograph (Kyoto, Japan) with the retention time of ethylene being 0.553 s.
Polyamine analysis
Polyamines analysis was carried out from leaves as described by Németh et al. (2002). 200 mg plant sample was ground in liquid nitrogen, extracted with 2 × 1 ml 0.2 M ice cold perchloric acid, put on ice for 20 min, then centrifuged at 4 °C for 20 min (10,000×g). In the case of free PA (polyamine) fraction, a dansyl-chloride derivative was prepared from 100 μl supernatant (Smith and Davies 1985). Dansylated polyamines were measured in a WATERS W 2690 (Milford, USA) HPLC instrument using acetonitrile (Sigma-Aldrich, Saint Louis, USA) carrier.
qPCR analysis
Primers were designed for the sequences of the genes to be examined, the main parameters of which are shown in Table S2. The reactions were performed using Corbett RG-6000 real time PCR equipment (Qiagen, Hilden, Germany). The reaction mixture contained (in 20 µl final volume): 10 µl 2×ABsolute qPCR SYBR Green Mix (ThermoFisher Scientific, Waltham, USA), 1.75 µl/primer (70 nM), 1 µl cDNA (diluted 1/50). For the determination of relative expression levels, the Comparative Quantification method was used supplied as part of Rotor-Gene Q Series Software 2.3.1. (Warton et al. 2004; McCurdy et al. 2008). N. benthamiana glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used as an endogenous control.
Microscopic analysis
The detection of FvSAMS::sGFP, the FvSAMDC::sGFP, the FvSPDS::sGFP and the 35Spro:sGFP fusion proteins was performed by Leica TCS SP8 confocal laser scanning microscope and Leica/Leitz DMRB 301-371.010 fluorescence stereo microscope (Leica, Wetzlar, Germany). The leaves were examined without fixation with 1:1 87% (v/v) glycerol:bidistilled water being used for covering. For labelling the endoplasmic reticulum CellLight™ ER-RFP, BacMam 2.0 reagent was used (ThermoFisher Scientific, Waltham, USA). In contrast to the manufacturer’s protocol with regard to the cell suspension, the CellLight™ ER-RFP was reinfiltrated into the living tissue (100-time dilution) followed by incubation at room temperature for 16 h before the fixation of the tissue [4% (w/v) paraformaldehyde, 0.1% (v/v) Tween 20, 0.1% (v/v) Triton X-100, 0.15 M NaCl, 10 mM sodium phosphate buffer pH 7.5]. The LAS AF Lite 3.3.10134.0 software was applied for the processing of the pictures. ImageJ 1.51j8 software was used for determining the relative fluorescence of sGFP (Schneider et al. 2012).
Statistical analysis
The results are the mean values of at least three measurements and were statistically evaluated using the standard deviation and ANOVA methods.
Results
Short-term salt stress and relative chlorophyll content
We examined the stress response of leaf discs of FvSAMS, FvSAMDC and WT N. benthamiana to short-term salt stress (Fig. 1a). We observed that the WT leaf explants died 48 h after being placed on MS medium containing 300 mM NaCl. At the same time, the death of the leaf tissues of FvSAMS and FvSAMDC lines occurred later, 96 h after salt exposure. The relative chlorophyll content of the leaf discs shown in Fig. 1a was quantified and presented in Fig. 1b. Chlorophyll content measured in four sampling times (24, 48, 72 and 96 h after treatment) also supports that there is a difference in the rate of chlorophyll degradation between the leaf discs of WT and transgenic FvSAMS, FvSAMDC lines in response to 300 mM NaCl stress. In the WT leaf explants the chlorophyll content decreased to ~ 15% as early as 48 h after the treatment, while the average value of FvSAMS and FvSAMDC transgenic leaf discs approached this level only in the 96th h.
Effect of 300 mM NaCl on the survival of Nicotiana benthamiana leaf discs. Young leaf discs (from the top to 4th node) were used 50 days after germination. Bars indicate 1 cm (a). Relative chlorophyll content of 300 mM NaCl stressed leaf discs, the control chlorophyll and chlorophyll-free samples were used for calibration (b)
Growth vigour, biomass production and chlorophyll content
The biomass of the plant is a good indication of its response to stress. We observed that both FvSAMS and FvSAMDC lines produced higher shoot biomass and shoot length than the non-transformed ones both in control and 10 mM NaCl stress conditions (Fig. 2a–c).
Visible effect of overexpression of FvSAMS and FvSAMDC on shoot and root biomass production in control (1–20) and 10 mM NaCl stress conditions (21–40). The numbers from 1 to 4 and 21 to 24 indicate the wild type, while 5 to 20 and 25 to 40 mark different transgenic lines. The same number refers to the same plant. Bars indicate 1 cm. Photographs were taken 85 days after germination (a). Dry weight of shoots (b). Results of shoot length measurements (c). Data are the means of six replicates (SD ±). Results of chlorophyll a and b content measurements (d). Data are the means of six (b) twelve (c) and three (d) replicates (SD ±), respectively. Three asterisks represent significant differences at P < 0.001 and one asterisk represents significant differences at P < 0.05
The chlorophyll content of plants demonstrates their physiological status, therefore we investigated the effect of long term 10 mM NaCl stress on the total chlorophyll content of transgenic FvSAMS and FvSAMDC lines and wild type plants. The results of chlorophyll a and b content measurements in Fig. 2d shows that the FvSAMS and FvSAMDC lines contained more chlorophyll than the WT ones both under stress-free and stressed conditions.
H2O2, O2− content and electrolyte leakage
The accumulation of ROSs (Reactive Oxygen Species), similarly to that of H2O2 and O2− is in direct proportion with the stress levels of cells, therefore we investigated the H2O2 and O2− contents of tissues by means of histochemical staining as well as quantitative tests. The histochemical staining indicated that the presence of H2O2 could hardly be seen in the stress-free leaves of either the WT or FvSAMS and FvSAMDC lines. More intense coloration of the salt stressed WT leaves corresponds to higher concentrations of H2O2 than in the FvSAMS and FvSAMDC lines (Fig. 3a, left panel). Similarly, to H2O2 more intense coloration appeared in the WT leaves indicating that there is a higher amount of O2− in the non-transformed plants than in the FvSAMS and FvSAMDC lines (Fig. 3a, right panel). Furthermore, salt treated leaves of FvSAMS and FvSAMDC lines showed low H2O2 contents compared to the WTs (Fig. 3b).
The measure of electrolyte leakage gives information about the status of the membranes. Therefore, we examined the electrolyte leakage of the leaf discs both under control and salt stress conditions (Fig. 3c). We detected lower rates of electrolyte leakage in both FvSAMS and FvSAMDC lines both under control and stress conditions than in the WTs.
Proline and lignin content
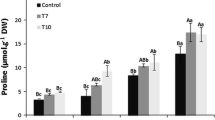
Proline accumulation is also considered to be a good indicator of stress response. There was no change of proline content in control condition. Salt stress increased (4.3 fold) the proline accumulation only in FvSAMDC lines (Fig. 4a).
Lignin is known to accumulate between the primary and secondary cell wall to ensure the strength of cell wall during plant development. A firm cell wall provides the cells with increased protection against biotic and abiotic stresses. We also measured the lignin content of the stem under both control and salt stress conditions (Fig. 4b). Under the control conditions more lignin was found in the shoots of FvSAMS and FvSAMDC lines than in WTs. The salt stress caused a rise in the lignin content of the WT and FvSAMS lines, but a significant decrease was found in the FvSAMDC genotypes.
Ethylene and polyamine content
Since ethylene is implicated in numerous stress responses, we investigated the effect of overproducing FvSAMS and FvSAMDC on the levels of ethylene both under control and stress conditions. In the FVSAMS lines there was no significant change in ethylene production either in control or salt stress conditions. The FvSAMDC lines did not produce significantly higher levels of ethylene under control conditions, but the salt stress induced a substantial rise in the ethylene production in the FvSAMDC transgenic plants than in WTs (Fig. 4c).
In addition to ethylene, we examined the free polyamine content of the transgenic FvSAMS and FvSAMDC lines and WT plants both under control and salinity conditions (Fig. 5). We detected significantly lower levels of putrescine (Put) in response to stress in the FvSAMS lines. However, the amount of Put was found to be significantly higher only in the FvSAMDC lines both under the control and stress conditions. In the FvSAMS lines significantly higher levels of spermidine (Spd) and spermine (Spm) were only detected in response to stress, while the amount of Spd and Spm was found to be significantly higher both under control and stress conditions in the FvSAMDC lines. In the FvSAMS lines significantly higher levels of total free polyamine content was only detected in response to stress, while the total free polyamine content was significantly higher in the FvSAMDC lines in response to both control and stress conditions.
Relative expression measurement
We analyzed the expression of genes participating in ethylene (SAM synthase—SAMS; ACC synthase—ACS) and polyamine biosynthesis (SAM synthase—SAMS; SAM decarboxylase—SAMDC; spermidine synthase—SPDS; spermine synthase—SPMS), the recycling of SAM (S-adenosyl-l-homocysteine hydrolase—SAHH) and the production of two secondary metabolites such as lignin (Cinnamyl-alcohol dehydrogenase—CAD) and nicotine (Putrescine N-methyltransferase—PMT) (Fig. 6). The relative expression rate of SAMS, SAMDC and SAHH was significantly higher in the FvSAMS and FvSAMDC lines than in the WTs both under control and salt stress conditions. The transcription rate of ACS was found to be lower in the FvSAMS lines under non-stressed condition and it was lower in response to salt stress as well. At the same time, FvSAMDC lines displayed higher levels of ACS expression than in the WTs under stress condition. The transcription rate of SPDS did not show significant difference between the WTs and the FvSAMS, FvSAMDC lines in stress-free environment, while it increased both in the FvSAMS and FvSAMDC lines in response to salt treatment. The relative expression rate of PMT was lower in the FvSAMS lines in the control environment and higher in the FvSAMDC lines than in the WTs. Significantly higher rate of PMT expression was measured in response to stress in both the FvSAMS and FvSAMDC lines. The expression levels of CAD were higher in almost all transgenic lines in control condition and all transgenics under salt stress than in the WTs.
The summary of the measurements in control and 10 mM NaCl stress conditions is shown in Table S1.
Subcellular localization of FvSAMS::sGFP and FvSAMDC::sGFP
We carried out transient expression investigations with the FvSAMS::sGFP, FvSAMDC::sGFP, FvSPDS::sGFP (F. vesca L. cv. Rügen spermidine synthase) and 35Spro:sGFP constructs. Transient expression of FvSAMS::sGFP, FvSAMDC::sGFP and 35Spro:sGFP fusion proteins displayed nuclear and cytoplasmic localization, while FvSPDS::sGFP showed only cytoplasmic distribution in epidermal cells (Fig. 7). We noticed that each of the three fusion proteins (FvSAMS::sGFP, FvSAMDC::sGFP and 35Spro:sGFP) showed endoplasmic reticulum (ER) signals during their transient expression (Fig. S1), therefore we used ER labelling with the application of CellLight™ ER-RFP (Fig. S2). The FvSAMS::sGFP and FvSAMDC::sGFP showed weaker ER signals than the 35Spro:sGFP construct. The fixation of the leaf tissue was carried out following the ER labelling, which resulted in the sGFP signal becoming less pronounced and therefore allowed us to observe the distinctive nuclear and nucleolar signals of FvSAMS::sGFP, which were detectable in the stable transformants as well (Fig. 8a, b). In the stable transformants, the FvSAMS::sGFP did not show homogeneous cytoplasmic distribution, but it accumulated in the outermost regions of the cytoplasm near the plasmalemma and plastid membrane in palisade parenchyma cells as it is visible in Fig. 8c and d. The distribution of FvSAMS::sGFP and FvSAMDC::sGFP fusion proteins in the stable transformants both under control and saline conditions is illustrated in Fig. 9a. We observed differences in the ratios of nuclear to cytoplasmic distribution under stress-free and stressed conditions. The nuclear signal of FvSAMS::sGFP was more intense in the control environment, and the cytoplasmic expression became stronger in response to salt stress (Fig. 9b, upper part). In the case of FvSAMDC::sGFP the amount of fusion proteins increased substantially in response to salt stress both in the nucleus and the cytoplasm (Fig. 9b, lower part). At the same time it could be observed that the FvSAMDC::sGFP showed distinctive nuclear, cytoplasmic and nuclear-cytoplasmic localization concurrently (Fig. 10a). We also noticed that in the case of FvSAMS lines the abundance of the FvSAMS::sGFP fusion protein changed depending on the position of the leaves in the plant. In the leaves closest to the apex the sGFP protein could be detected in almost all of their cells while moving downwards the number of sGFP expressing cells decreased, finally leaving only the guard cells of the stomata giving off the sGFP signal (Fig. 10b, ci, ii). Therefore, we determined the amount of sGFP mRNA in the leaves from the top to 4th node and from the 5th to 7th nodes in the FvSAMS lines. Despite the fact that the detectability of sGFP protein was higher in the upper leaves the amount of mRNA showed the opposite trend with the lower leaves containing ~ 2–5-times more mRNA than the upper ones (Fig. 10d). Similar phenomenon was not observed in the FvSAMDC lines.
FvSAMS::sGFP signals in the cell nucleus and nucleolus of epidermal cells with transient expression (a) and in the palisade parenchyma cells of the stable transformant plants in 3D (b). The cytoplasmic (near to the plasmalemma) and plastid membrane localization of FvSAMS::sGFP in palisade parenchyma cells (c) and these sGFP signals in 3D (d). The iv–iv’ and v–v’ refer to the same nucleus. Bars indicate 10 µm (a, c, d) and 20 µm (b). Arrows indicate plastid membrane (a) and nucleolus (c, d) localization of FvSAMS::sGFP
Subcellular localization of FvSAMS::sGFP and FvSAMDC::sGFP in palisade parenchyma cells of stable transformants in control and salt stress conditions. Bars indicate 10 µm (a). Relative fluorescence intensity of FvSAMS::sGFP and FvSAMDC::sGFP fusion protein in the nucleus and cytoplasm under control and salinity conditions (a. u.: arbitrary unit). Data are the means of ten replicates (SD ±). Three asterisks represent significant differences at P < 0.001 (b)
Different subcellular localization of FvSAMDC::sGFP in palisade parenchyma cells. Bars indicate 10 µm (a). Detection of FvSAMS::sGFP expression in different leaf levels. Bars indicate 30 µm (b). Detecting the expression of FvSAMS::sGFP protein in the epidermal cells, palisade parenchyma cells and guard cells of the stomata in the 1st (i) to 5th (ii) leaves from above (c). Relative expression of FvSAMS from leaves at different levels. Data are means of four replicates (SD ±). Three asterisks represent significant differences at P < 0.001 (d)
Bioinformatic analysis
To further investigate the nuclear, nucleolar and cytoplasmic (near to the plasmalemma), plastid membrane localization of FvSAMS::sGFP and the nuclear, homogenous cytoplasmic localization of FvSAMDC::sGFP, the amino acid of FvSAMS and FvSAMDC sequences were examined by means of various bioinformatic applications. We searched for nuclear localization signals (NLSs), the nuclear export signals (NESs), the nucleolar localization signals (NoLSs) and potential post-translational modification sites in these sequences. Figure 11a shows the absence of a cysteine residue in the enzyme SAMS, which plays an important role in the inhibition of the SAMS by S-nitrosylation. Furthermore, Fig. 11b shows the recognition sequence involved in the inhibition of enzyme SAMS by Protein kinase C. Both FvSAMS and FvSAMDC contain putative bipartite nuclear localization signal and nuclear export signal sequences, but FvSAMS contained no predicted nucleolar localization signals. The predicted post-translational modification sites of FvSAMS include Ser-38 and Thr-112 for phosphorylation, Lys-335, 360 and 364 for acetylation, Cys-20 and 31 for S-nitrosylation, Lys-39 and 334 for SUMOylation. Potential sites of post-translational modifications of FvSAMDC based on Ser-25, 37, 39, 203, 204, 212, 316 and 317 for phosphorylation, Lys-199 for acetylation, Cys-312 for S-nitrosylation and Lys-334 for SUMOylation (Fig. 11c, d).
Bioinformatic analysis of FvSAMS and FvSAMDC. S-nitrosylation region of AtSAMS1 (NP_171751.1) proteins (black rectangular) and the lack of cystein (C) necessary for S-nitrosylation in FvSAMS (XP_004288342.1), SlSAMS1 (NP_001234425.1) and StSAMS1 (NP_001275609.1) (red rectangle). Dots mark amino acids identical to the sequence of AtSAMS1 (a). Predicted phosphorylation site of Thr-112 and the possible recognition sequence of Protein kinase C (red rectangle) (b). Bioinformatic analysis of the FvSAMS (c) and FvSAMDC (d) amino acid sequences. Colour code: predicted bipartite NLS (nuclear localization signal) (red letters), predicted S-nitrosylation Sites (highlighted in yellow, in areas of overlap not bold italics), predicted Phosphorylation Sites (highlighted in turquoise), predicted Acetylation Sites (blue letters), predicted SUMOylation (highlighted in green), overlap between predicted NLSs and the post-translational modification sites (star). In the case of FvSAMS the N— (blue arrow), Central-(white arrow) and the C-terminal domains (red arrow) are indicated according to the UniProtKB—Q96551
Discussion
In the present study we performed a comparative investigation in an attempt to find out how the overexpression of FvSAMS and FvSAMDC affects the various physiological parameters of the plants in response to salt stress. We found that the overproduction of FvSAMS and FvSAMDC decreased chlorophyll degradation in 300 mM NaCl stress (Fig. 1) and increased the biomass in response to 10 mM NaCl stress (Fig. 2), in agreement with earlier findings by others (Roy and Wu 2002; Wi et al. 2006; Qi et al. 2010; Gong et al. 2014; Kim et al. 2015; Ma et al. 2017). However, we found significant differences in the above mentioned parameters even under control conditions compared to the WT plants.
The chloroplasts are the main sites of the production of ROS, including superoxide anions, hydrogen peroxide, hydroxyl radical and singlet oxygen (Mignolet-Spruyt et al. 2016). ROS homeostasis is altered in response to stress. When present at high concentrations, ROS can damage the cell and at low concentrations they act as signals in recognising stress and triggering a response. Excessive accumulation of ROS causes oxidative stress, which leads to lipid peroxidation and membrane structure damage, leading eventually to cell death (Biswas and Mano 2015). Increased membrane stability accounts for significantly lower rates of electrolyte leakage and H2O2, O2− in both FvSAMS and FvSAMDC lines both under control and stress conditions than in the WTs (Fig. 3). Lower electrolyte loss was measured in SlSAMS, MfSAMS1 overproducing tomato and tobacco plants in salt stress conditions (Gong et al. 2014; Guo et al. 2014) and in heat-stressed tomato plants overexpressing yeast SAMDC (Cheng et al. 2009) than in the wild types. Significantly lower levels of ROS accumulation could be detected in SlSAMS and BvM14-SAMS2 overproducing tomato, A. thaliana (Gong et al. 2014; Ma et al. 2017) and in CaSAMDC overexpressing A. thaliana plants (Wi et al. 2014). Several previous publications reported that the overproduction of SAMS and SAMDC increased the activity of the ROS scavenging enzymes through increased polyamine biosynthesis (Wi et al. 2006, 2014; Gong et al. 2014; Guo et al. 2014; Ma et al. 2017). There are two biochemical pathways by which polyamines can exert their control over the homeostasis of ROS. One of the pathways leads to the inhibition of the auto-oxidation of metals, which decreases the electron supply required for the generation of ROS (Shi et al. 2010). The other pathway directly increases the activity of enzymes in the antioxidant system (Shi et al. 2010; Mostofa et al. 2014), which presumably leads to the reduced quantity of H2O2 and O2− in the transgenic FvSAMS and FvSAMDC lines.
Besides the ROS, the amount of accumulating proline in the cell is also a good indicator of the level of stress the plants are exposed to. The elevated proline levels have numerous positive impacts on the abiotic stress tolerance of plants, with proline accumulation being directly proportional to the levels of polyamines, in spite of the fact that they share a common precursor (glutamic acid) (Simon-Sarkadi et al. 2006; Wen et al. 2011; Cvikrová et al. 2012). Higher levels of total polyamine measured in our FvSAMS-expressing lines showed no positive correlations with the proline content detected. In contrast to our results, higher proline levels were found in A. thaliana plants overproducing BvM14-SAMS2 in response to salt stress than in WT plants. The overexpression of FvSAMDC increased the levels of proline in response to salt stress (Fig. 4a), which is in agreement with the higher levels of total polyamine measured in FvSAMDC lines.
Since lignin is one of the main metabolic consumers of SAM, we investigated how ‘SAM flux’ and ‘SAM deficiency’ influence the lignin content of plants under control conditions and in response to salt stress. Lignin is deposited between the primary and secondary cell walls during plant development and thereby increases the strength of the stem, which, in turn, improves the tolerance of plants against biotic and abiotic stresses. Accelerated lignification was observed in the roots of water-deficient sorghum, salt stressed soybean and salt stressed maize plants (Azaizeh and Steudle 1991; Cruz et al. 1992; Neves et al. 2010). Sánchez-Aguayo et al. (2004) concluded that the elevated activity of SAM in salt stressed tomato plants is commensurate with the increased amount of lignin deposited in vascular tissues. According to our result, both ‘SAM flux’ and ‘SAM deficiency’ increased the rate of lignification in the transgenic FvSAMS and FvSAMDC lines under control conditions. However, in response to salt stress ‘SAM deficiency’ substantially reduced, while ‘SAM flux’ substantially increased lignification, which could have directly increased the tolerance of FvSAMS lines against salt stress (Fig. 4b).
Since SAM is a precursor molecule of ethylene biosynthesis, we also investigated how the overexpression of FvSAMS and FvSAMDC influenced the amount of ethylene under control and stress conditions. Although the overproduction of FvSAMS had no influence on the release of ethylene, the overexpression of FvSAMDC upregulated ethylene production under stress conditions (Fig. 4c). In previous studies, down-regulated tobacco SAMDC activity did not cause any changes in ethylene production (Torrigiani et al. 2005), while the overexpression of AtSAMDC in A. thaliana plants reduced the amount of ethylene (Hu et al. 2006). Furthermore, neither the overexpression of FvSAMS, nor that of MfSAMS1 in tobacco plants resulted in any significant changes in the amount of ethylene produced (Guo et al. 2014), similarly to our results. However, the overexpression of SAMS increased the level of ethylene production in tomato and A. thaliana plants (Gong et al. 2014; Kim et al. 2015).
Polyamines are known to play an important role in osmotic regulations, membrane stability, and the removal of free radicals under abiotic stresses (Liu et al. 2007; Romero et al. 2018). In accordance with earlier studies, the overexpression of both FvSAMS and FvSAMDC was found to increase the total polyamine levels in response to stress (Roy and Wu 2002; Waie and Rajam 2003; Wi et al. 2006, 2014; Cheng et al. 2009; Peremarti et al. 2009; Gong et al. 2014; Ma et al. 2017). However, the overproduction of the FvSAMDC increased the total polyamine levels more substantially, while the “SAM surplus” caused a lesser degree of accumulation in the FvSAMS lines (Fig. 5). It follows therefore that in contrast to several earlier studies (Li et al. 2004; Nambeesan et al. 2012; Yu et al. 2016), no antagonistic effect between ethylene and polyamine biosynthesis was observed in our experimental system.
Based on the data measured in the FvSAMDC lines there appears to be a positive correlation between the free PA levels and the proline content as well as the amount of ethylene, while there is a negative correlation between the free PA levels and the lignin content when the plants are exposed to salt stress (Figs. 4c, 5).
The overexpression of FvSAMS and FvSAMDC increased the relative expression rates of genes SAMS, SAMDC, SPDS, SPMS, SAHH, PMT and CAD under stress condition (Fig. 6). However, the expression rates of ACS decreased in the FvSAMS but increased in the FvSAMDC lines under stress conditions, which was also supported by the ethylene production (Figs. 4c, 6). The relatively high levels of expression of CAD in response to stress in the FvSAMDC lines showed no positive correlation with lignin contents (Figs. 4b, 6), according to our assumption the lower lignin content measured in the FvSAMDC lines can induce the higher CAD expression in the case of salt stress in order to increase the lignin production. In earlier reports the overexpression of SAMS increased the relative expression rates of the genes SAMS, SAMDC, ACS, SPDS and SPMS in tomato, Medicago sativa and A. thaliana (Gong et al. 2014; Guo et al. 2014; Kim et al. 2015) and the overproduction of SAMDC increased the relative expression levels of SAMDC and decreased those of ACS in tobacco and A. thaliana (Wi et al. 2006, 2014).
In our opinion the prolonged survival and increased salt tolerance of the FvSAMDC lines can be attributed to the higher level of polyamines, while in the FvSAMS lines elevated lignin and polyamines contents together were among the contributing factors.
We investigated the subcellular localization of the proteins encoded by FvSAMS::sGFP and FvSAMDC::sGFP. The FvSAMS::sGFP exhibited nuclear, nucleolar and cytoplasmic (near to the plasmalemma), plastid membrane localization in stable transformants (Fig. 8c). Previous studies reported that the SAMS is considered to be an enzyme with cytosolic localization in plants (Schröder et al. 1997; Ravanel et al. 1998; Hanson and Roje 2001), whereas in mammals it was found to show nucleocytoplasmic distribution (Reytor et al. 2009). Reytor et al. (2009) reported that the nuclear accumulation of MAT (methionine adenosyltransferase = SAMS in plant) showed positive correlations with the trimethylation of the histone H3K27. FvSAMDC::sGFP showed weak nuclear and homogeneous cytoplasmic distribution (Fig. 9a). In former studies SAMDC activities were found in the chloroplasts, mitochondria and cytosol of plants (Yamanoha and Cohen 1985; Torrigiani et al. 1986), while only cytoplasmic localization was detected in animals (Gritli-Linde et al. 1995). Since we detected no chloroplast or mitochondrial sGFP signals, the amino acid sequence of FvSAMDC was examined bioinformatically. According to application, prediction of the probability of FvSAMDC having chloroplast, mitochondrial and other localization was found to be 7%, 9% and 80%, respectively.
In the case of both constructs, endoplasmic reticulum subcellular localization was observed during transient expression, but no similar signals were detected in the FvSAMS and FvSAMDC stable transformants (Figs. S1, S2). We assumed therefore that these signals could be interpreted as a response to the physical and biotic stresses caused by the infiltration procedure. This phenomenon cannot be adequately explained by our current data and requires further investigations, but both biotic and abiotic stresses can cause the accumulation of unfolded proteins, which interact with the specific sensor proteins of the ER membrane. The response to the unfolded proteins (UPR—unfolded protein response) is a conserved eukaryotic stress reaction, and it was the unfolded protein ER-associated degradation (ERAD) that we presumably observed (Walter and Ron 2011).
In our experiments the FvSAMS::sGFP protein was localized to the nucleolus, an observation which has not been reported previously (Fig. 8a, b). Reytor et al. (2009) found no nucleolar localization of the MAT proteins. With the exception of FC (Fibrillar Center) shown in Fig. 8a ii, the FvSAMS::sGFP is localized either in DFC (Dense Fibrillar Component) or in GC (Granular Component) or in both nucleolar components, which take part in the biogenesis of ribosomes. DFC contains the protein fibrillarin (Sirri et al. 2008), which plays a role in the processing of rRNA. Furthermore, an MTase region (Ado-Met-dependent methyltransferase) was identified in the N. benthamiana Fibrillarin2 protein (NbFib2), which contained SAM binding motives and localized to the nucleolus (Zheng et al. 2016). Therefore, the nucleolar localization of FvSAMS::sGFP suggests that it plays a role in ribosome synthesis. The fact that the FvSAMS sequences contain no nucleolar localization signals makes it probable that there is an intermediate interaction required for the nucleolar localization of FvSAMS::sGFP proteins, but this phenomenon requires further investigations. Previous studies reported that SAM is delivered into plastids by specific SAM transporters (Ferro et al. 2002; Palmieri et al. 2006), where SAM is required to methylation of DNA, RNA and proteins (Montasser-Kouhsari et al. 1978; Block et al. 2002). According to our results the FvSAMS::sGFP showed nuclear, nucleolar and cytoplasmic (near to the plasmalemma), plastid membrane distribution, therefore SAM is assumed to be produced in situ for numerous biochemical reactions (e.g. ribosome biosynthesis, methylation of DNA, RNA, proteins in nucleus and plastid; lignin biosynthesis).
Our results showed that in stable transformants the subcellular localization of the FvSAMS::sGFP protein was displaced in the direction of the cytoplasm (near to the plasmalemma and plastid membrane) from the nucleus in response to stress, while the quantity of FvSAMDC::sGFP protein showed a parallel increase in both cellular compartments (nucleus, cytoplasm) (Fig. 9b). Several nuclear proteins carry both NLS and NES signals, which control their transport between the cell nucleus and the cytoplasm. The presence of putative bipartite nuclear localization and nuclear export signals in both FvSAMS and FvSAMDC is in agreement with the nuclear and cytoplasmic localization of both enzymes, though the fictionality of these signals remain to be demonstrated experimentally.
We could randomly detect FvSAMDC::sGFP fusion proteins only in a few cases from several dozens of preparations, which can be due to the short half life time of the SAMDC (Pegg 1986), but it could be observed that the FvSAMDC::sGFP showed distinctive nuclear, cytoplasmic and nuclear-cytoplasmic localization concurrently in the stable transformants (Fig. 10a). The fact that it showed nuclear and cytoplasmic localization separately and also simultaneously can be explained by the differential autocatalytic activation of the proenzyme SAMDC in the different cellular compartments as well as by the unidirectional dislocation of the enzyme. However, we were not able to identify whether the direction of its movement is cytoplasm → nucleus or nucleus → cytoplasm. The dual localization of the FvSAMDC::sGFP is also supported by an earlier study (Belda-Palazón et al. 2012) in which it was described that the spermidine synthase showed nuclear and cytoplasmic localization, whereas the spermine synthase had only cytoplasmic localization. Belda-Palazón et al. (2012) also observed that in the presence of exogenic spermidine synthase, the cytoplasmic localization of spermine synthase showed a displacement towards the nucleus, which suggests that the synthesis of spermidine and spermine is a complex process. Since SAMDC produces dcSAM, which serves as a substrate for both the spermidine and spermine synthases by providing them with aminopropyl group, the synthesis of SAMDC in an inactive form, its strict transcriptional and translational regulation as well as its short half-life can account for its dual localization due to the complexity of the process.
We also observed that the amount of detectable FvSAMS::sGFP protein showed a decreasing tendency from the younger leaves towards the older ones but higher FvSAMS mRNA rates were measured in the older leaves in spite of the lower levels of detectable FvSAMS::sGFP protein. Finally, FvSAMS::sGFP fusion proteins were only detected in the guard cells of older leaves (Fig. 10b–d), therefore we suppose that the SAMS enzyme has an important role to play in the regulation of the stomatal functions, such as cellular respiration, photosynthetic gas exchange and evaporation. Similar pattern was obtained in tobacco plants overexpressing AtSAMS1 by Boerjan et al. (1994), namely that AtSAMS1 mRNA accumulation grew from top to bottom but the activity of AtSAMS1 enzyme showed a reverse correlation to the amount of mRNA. Later it was proven that AtSAMS1 is inhibited by nitric oxide (NO). Structural analysis of AtSAMS1 showed that the Cys-114 is flanked with amino acids promoting S-nitrosylation. We found that the amino acid sequence of SAMS contained arginine (R) at the site of Cys-114 (Fig. 11a). The inhibitory effect of S-Nitrosoglutathione (GSNO) drastically decreased when Cys-114 was replaced by arginine (Lindermayr et al. 2006). Earlier studies focused on the reduction in the activity of enzyme SAMS by S-nitrosylation (Avila et al. 1997; Lindermayr et al. 2006), while other publications discussed the phosphorylation of Thr-342 by Protein kinase C (PKC), which decreased the enzyme activity of SAMS (Pajares et al. 1994) as well as the predicted phosphorylation of Thr, Ser, Thr-112 and Ser-271 in FvSAMS (Fig. 11b), which is equivalent to the S/TXK/R (Woodgett et al. 1986) and TXRX (Pajares et al. 1994) recognition sequences preferred by PKC (in the case of FvSAMS the TKRP and SGK). Therefore, it is possible that we assume that the phosphorylation of Thr-112 and Ser-271 play a role in the inhibition of enzyme FvSAMS by PKC, but we currently do not have evidence for this possibility.
According to Reytor et al. (2009) the subcellular localization of the enzyme SAMS is determined by the structure of its C-terminal domain, which makes the involvement of Lys-335, 360, 364 acetylation and Lys-387 SUMOylation in the regulation of subcellular localization of FvSAMS probable (Fig. 11c). The results of earlier studies support this hypothesis since the SUMOylation of NLS of the human enzyme SAE resulted in its nuclear accumulation (Truong et al. 2012) and the acetylation of the NLS of the human RhoA GEF Net1A increased its cytoplasmic accumulation (Song et al. 2015). We have no data available with regard to the post-translational modifications determining the subcellular localization of the enzyme FvSAMDC but based on predicted data the phosphorylation, acetylation, S-nitrosylation and SUMOylation of the NLSs can play a role in the process (Fig. 11d), since numerous published data provided evidence supporting the role of NLS phosphorylation, acetylation, S-nitrosylation, SUMOylation in the subcellular localization of enzymes (Harreman et al. 2004; Hara et al. 2005; Truong et al. 2012; Song et al. 2015).
References
Ábrahám E, Hourton-Cabassa C, Szabados L (2010) Methods for determination of proline in plants. In: Sunkar R (ed) Plant stress tolerance, methods in molecular biology. Humana Press, Totowa, pp 317–331
Avila MA, Mingorance J, Martínez-Chantar ML, Casado M, Martín-Sanz P, Bosca L, Mato JM (1997) Regulation of rat liver S-adenosylmethionine synthetase during septic shock: role of nitric oxide. Hepatology 25:391–396
Azaizeh H, Steudle E (1991) Effects of salinity on water transport of excised maize (Zea mays L.) roots. Plant Physiol 97:1136–1145
Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A (2012) Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7:e46907
Bennett EM, Ekstrom JL, Pegg AE, Ealick SE (2002) Monomeric S-adenosylmethionine decarboxylase from plants provides an alternative to putrescine stimulation. Biochemistry 41:14509–14517
Biswas MS, Mano J (2015) Lipid peroxide-derived short-chain carbonyls mediate H2O2-induced and NaCL-induced programmed cell death in plants. Plant Physiol 168:885–898
Block MA, Tewari AK, Albrieux C, Marechal E, Joyard J (2002) The plant S-adenosyl-l-methionine: Mg-protoporphyrin IX methyltransferase is located in both envelope and thylakoid chloroplast membranes. Eur J Biochem 269:240–248
Boerjan W, Bauw G, Montagu MV, Inzé D (1994) Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6:1401–1414
Cantoni GL (1975) Biological methylation: selected aspects. Annu Rev Biochem 44:435–451
Cheng L, Zou Y, Ding S, Zhang J, Yu X, Cao J, Lu G (2009) Polyamine accumulation in transgenic tomato enhances the tolerance to high temperature stress. J Integr Plant Biol 51:489–499
Chinnusamy V, Zhu J, Zhu JK (2006) Salt stress signaling and mechanisms of plant salt tolerance. Genet Eng 27:141–177
Clemente T (2006) Nicotiana (Nicotiana tabacum, Nicotiana benthamiana). Methods Mol Biol 343:143–154
Cruz RT, Jordan WR, Drew MC (1992) Structural changes and associated reduction of hydraulic conductance in roots of Sorghum bicolor L. following exposure to water deficit. Plant Physiol 99:203–212
Cvikrová M, Gemperlová L, Dobrá J, Martincová O, Prásil IT, Gubis J, Vanková R (2012) Effect of heat stress on polyamine metabolism in proline-over-producing tobacco plants. Plant Sci 182:49–58
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP, and related tools. Nat Protoc 2:953–971
Ferro M, Salvi D, Riviere-Rolland H, Vermat T, Seigneurin-Berny D, Grunwald D, Garin J, Joyard J, Rolland N (2002) Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters. Proc Natl Acad Sci USA 99:11487–11492
Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29:243–249
Franceschetti M, Hanfrey C, Scaramagli S, Torrigiani P, Bagni N, Burtin D, Michael AJ (2001) Characterization of monocot and dicot plant S-adenosyl-l-methionine decarboxylase gene families including identification in the mRNA of a highly conserved pair of upstream overlapping open reading frames. Biochem J 353:403–409
Ge C, Cui X, Wang Y, Hu Y, Fu Z, Zhang D, Cheng Z, Li J (2006) BUD2, encoding an S-adenosylmethionine decarboxylase, is required for Arabidopsis growth and development. Cell Res 16:446–456
Gnad F, Gunawardena J, Mann M (2011) PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res 39:253–260
Gong B, Xiu L, Vandenlangenberg KM, Dan W, Shasha S, Min W, Yan L, Fengujan Y, Qinghua S, Xiufeng W (2014) Overexpression of S-adenosyl-l-methionine synthetase increased tomato tolerance to alkali stress trough polyamine metabolism. Plant Biotechnol J 12:694–708
Gritli-Linde A, Holm I, Linde A (1995) Localization of S-adenosylmethionine decarboxylase in murine tissues by immunohistochemistry. Eur J Oral Sci 103:133–140
Guo Z, Tan J, Zhuo C, Wang C, Xiang B, Wang Z (2014) Abscisic acid, H2O2 and nitric oxide interactions mediated cold-induced S-adenosylmethionine synthetase in Medicago sativa subsp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol J 12:601–612
Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Phys 52:119–137
Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat Cell Biol 7:665–674
Harreman MT, Kline TM, Milford HG, Harben MB, Hode AE, Corbett AH (2004) Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J Biol Chem 279:20613–20621
Hu WW, Gong H, Pua EC (2005) The pivotal roles of plant S-adenosylmethionine decarboxylase 5’ untranslated leader sequence in regulation of gene expression at the transcriptional and post-transcriptional levels. Plant Physiol 138:276–286
Hu WW, Gong H, Pua EC (2006) Modulation of SAMDC expression in Arabidopsis thaliana alters in vitro shoot organogenesis. Physiol Plant 128:740–750
Jang SJ, Wi SJ, Choi YJ, An G, Park KY (2012) Increased polyamine biosynthesis enhances stress tolerance by preventing the accumulation of reactive oxygen species: T-DNA mutational analysis of Oryza sativa lysine decarboxylase-like protein 1. Mol Cells 34:251–262
Kamal AH, Cho K, Kim DE, Uozumi N, Chung KY, Lee SY, Choi JS, Cho SW, Shin CS, Woo SH (2012) Changes in physiology and protein abundance in salt-stressed wheat chloroplasts. Mol Biol Rep 39:9059–9074
Kim SH, Kim SH, Palaniyandi SA, Yang SH, Suh JW (2015) Expression of potato S-adenosyl-l-methionine synthase (SbSAMS) gene altered developmental characteristics and stress responses in transgenic Arabidopsis plants. Plant Physiol Biochem 87:84–91
Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106:10171–10176
Kumar D, Yusuf MA, Singh P, Sardar M, Sarin NB (2014) Histochemical detection of superoxide and H2O2 accumulation in Brassica juncea seedlings. Bio-Protoc 4:1108
Kusano T, Suzuki H (2015) Polyamine homeostasis in plants: the role(s) of evolutionarily conserved upstream ORFs. In: Kusano T, Suzuki H (eds) Polyamines: a universal molecular nexus for growth, survival, and specialized metabolism. Springer, Tokyo, pp 111–118
Kusano T, Berberich T, Tateda C, Takahashi Y (2008) Polyamines: essential factors for growth and survival. Planta 228:367–381
La Cour T, Kiemer L, Mølgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17:527–536
Langebartels C, Kerner K, Leonardi S, Schraudner M, Trost M, Heller W, Sandermann H Jr (1991) Biochemical plant responses to ozone. I. Differential induction of polyamine and ethylene biosynthesis of tobacco. Plant Physiol 95:882–889
Li X (2011) Infiltration of Nicotiana benthamiana protocol for transient expression via Agrobacterium. Bio-Protoc 101:e95
Li C, Jiao J, Wang G (2004) The important roles of reactive oxygen species in the relationship between ethylene and polyamines in leaves of spring wheat seedlings under root osmotic stress. Plant Sci 166:303–315
Lindermayr C, Saalbach G, Bahnweg G, Durner J (2006) Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J Biol Chem 281:4285–4291
Liu JH, Kitashiba H, Wang J, Ban Y, Moriguchi T (2007) Polyamines and their ability to provide environmental stress tolerance to plants. Plant Biotechnol 24:117–126
Liu JH, Inoue H, Moriguchi T (2008) Salt stress-mediated changes in free polyamine titers and expression of genes responsible for polyamine biosynthesis of apple in vitro shoots. Environ Exp Bot 62:28–35
Ma C, Wang Y, Gum D, Nan J, Chen S, Li H (2017) Overexpression of S-adenosyl-l-methionine synthetase 2 from sugar beet M14 increased Arabidopsis tolerance to salt and oxidative stress. Int J Mol Sci 18:847
Mahajan S, Tuteja N (2005) Cold, salinity and drought stresses: an overview. Arch Biochem Biophys 444:139–158
McCurdy RD, McGrath JJ, Mackay-Sim A (2008) Validation of the comparative quantification method of real-time PCR analysis and a cautionary tale of housekeeping gene selection. Gene Ther Mol Biol 12:15–24
Mellidou I, Moschou PN, Ioannidis NE, Pankou C, Gėmes K, Valassakis C, Andronis EA, Beris D, Haralampidis K, Roussis A, Karamanoli A, Matsi T, Kotzabasis K, Constantinidou HI, Roubelakis-Angelakis KA (2016) Silencing S-adenosyl-l-methionine decarboxylase (SAMDC) in Nicotiana tabacum points at a polyamine-dependent trade-off between growth and tolerance responses. Front Plant Sci 7:379
Merali S, Clarkson AB Jr (2004) S-adenosylmethionine and pneumocystis. FEMS Microbiol Lett 237:179–186
Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J (2016) Spreading the news: subcellular and organellar reactive oxygen species production and signalling. J Exp Bot 67:3831–3844
Montasser-Kouhsari S, Keith G, Weil GH (1978) Methylation of yeast tRNAPhe by enzymes from cytoplasm, chloroplasts and mitochondria of Phaseolus vulgaris. Biochim Biophys Acta 521:576–583
Moreira-Vilar FC, de Cassia Siqueira-Soares R, Finger-Teixeira A, de Oliveira DM, Ferro AP, da Rocha GJ, Ferrarese MD, dos Santos WD, Ferrarese-Filho O (2014) The acetyl bromide method is faster, simpler and presents best recovery of lignin in different herbaceous tissues than Klason and thioglycolic acid methods. PLoS ONE 9:e110000
Mostofa MG, Yoshida N, Fujita M (2014) Spermidine pretreatment enhances heat tolerance in rice seedlings through modulating antioxidative and glyoxalase systems. Plant Growth Regul 73:31–44
Müller M, Munné-Bosch S (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol 169:32–41
Nambeesan S, AbuQamar S, Laluk K, Mattoo AK, Mickelbart MV, Ferruzzi MG, Mengiste T, Handa AK (2012) Polyamines attenuate ethylene-mediated defense responses to abrogate resistance to Botrytis cinerea in tomato. Plant Physiol 158:1034–1045
Németh M, Janda T, Horváth E, Páldi E, Szalai G (2002) Exogenous salicylic acid increases polyamine content but may decrease drought tolerance in maize. Plant Sci 162:569–574
Neves GYS, Marchiosi R, Ferrarese MLL, Siqueira-Soares RC, Ferrarese-Filho O (2010) Root growth inhibition and lignification induced by salt stress in soybean. J Agron Crop Sci 196:467–473
Pajares MA, Duran C, Corrales F, Mato JM (1994) Protein kinase C phosphorylation of rat liver S-adenosylmethionine synthetase: dissociation and production of an active monomer. Biochem J 303:949–955
Palmieri L, Arrigoni R, Blanco E, Carrari F, Zanor MI, Studart-Guimaraes C, Fernie AR, Palmieri F (2006) Molecular identification of an Arabidopsis S-Adenosylmethionine transporter. analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol 142:855–865
Pegg AE (1986) Recent advances in the biochemistry of polyamines of eukaryotes. Biochem J 234:249–262
Peng J, Li Z, Wen X, Li W, Shi H, Yang L, Zhu H, Guo H (2014) Salt-induced stabilization of EIN3/EIL1 confers salinity tolerance by deterring ROS accumulation in Arabidopsis. PLoS Genet 10:e1004664
Peremarti A, Bassie L, Christou P, Capell T (2009) Spermine facilitates recovery from drought but does not confer drought tolerance in transgenic rice plants expressing Datura stramonium S-adenosylmethionine decarboxylase. Plant Mol Biol 70:253–264
Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim Biophys Acta 975:384–394
Pulla RK, Kim YJ, Parvin S, Shim JS, Lee JH, Kim YJ, In JG, Senthil KS, Yang DC (2009) Isolation of S-adenosyl-l-methionine synthetase gene from Panax ginseng C. A. meyer and analysis of its response to abiotic stresses. Physiol Mol Biol Plants 15:267–275
Qi YC, Wang FF, Zhang H, Liu WQ (2010) Overexpression of Suadea salsa S-adenosylmethionine synthetase gene promotes salt tolerance in transgenic tobacco. Acta Physiol Plant 32:263–269
Ravanel S, Gakière B, Job D, Douce R (1998) The specific features of methionine biosynthesis and metabolism in plants. Proc Natl Acad Sci USA 95:7805–7812
Reytor E, Pérez-Miguelsanz J, Alvarez L, Pérez-Sala D, Pajares MA (2009) Conformational signals in the C-terminal domain of methionine adenosyltransferase I/III determine its nucleocytoplasmic distribution. FASEB J 23:3347–3360
Rizhsky L, Hallak-Herr E, Van Breusegem F, Rachmilevich S, Barr JE, Rodermel S, Inzé D, Mittler R (2002) Double antisense plants lacking ascorbate peroxidase and catalase are less sensitive to oxidative stress than single antisense plants lacking ascorbate peroxidase or catalase. Plant J 32:329–342
Roeder S, Dreschler K, Wirtz M, Cristescu SM, van Harren FJ, Hell R, Piechulla B (2009) SAM levels, gene expression of SAM synthase, methionine synthase and ACC oxidase, and ethylene emission from N. suaveolens flowers. Plant Mol Biol 70:535–546
Romero FM, Maiale SJ, Rossi FR, Marina M, Ruíz OA (2018) Gárriz A (2018) Polyamine metabolism responses to biotic and abiotic stress. In: Alcazar R, Tiburcio AF (eds) Polyamines: methods and protocols, methods in molecular biology. Humana Press, New York, pp 37–49
Roy M, Wu R (2002) Overexpression of S-adenosylmethionine decarboxylase gene in rice increases polyamine level and enhances sodium chloride-stress tolerance. Plant Sci 163:987–992
Saha J, Brauer EK, Sengupta A, Popescu SC, Gupta K, Gupta B (2015) Polyamines as redox homeostasis regulators during salt stress in plants. Front Environ Sci 3:1–13
Sánchez-Aguayo I, Rodríguez-Galán JM, García R, Torreblanca J, Pardo JM (2004) Salt stress enhances xylem development and expression of S-adenosyl-l-methionine synthase in lignifying tissues of tomato plants. Planta 220:278–285
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Schröder G, Eichel J, Breining S, Schröder J (1997) Three differentially expressed S-adenosylmethionine synthetase from Catharantus roseus: molecular and functional characterization. Plant Mol Biol 33:211–222
Scott MS, Troshin PV, Barton GJ (2011) NoD: a Nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinform 12:317
Shen B, Li C, Tarczynski MC (2002) High free-methionine and decreased lignin content result from a mutation in the Arabidopsis S-adenosyl-l-methionine synthetase 3 gene. Plant J 29:371–380
Shen X, Wang Z, Song X, Xu J, Jiang C, Zhao Y, Ma C, Zhang H (2014) Transcriptomic profling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol Biol 86:303–317
Shi J, Fu XZ, Peng T, Huang XS, Fan QJ, Liu JH (2010) Spermine pretreatment confers dehydration tolerance of citrus in vitro plants via modulation of antioxidative capacity and stomatal response. Tree Physiol 30:914–922
Simon-Sarkadi L, Kocsy G, Várhegyi Á, Galiba G, de Ronde JA (2006) Effect of drought stress at supraoptimal temperature on polyamine concentrations in transgenic soybean with increased proline levels. Z Naturforsch C 61:833–839
Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (2008) Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129:13–31
Smith MA, Davies PJ (1985) Separation and quantitation of polyamines in plant tissue by high performance liquid chromatography of their dansyl derivatives. Plant Physiol 78:89–91
Song EH, Oh W, Ulu A, Carr HS, Zuo Y, Frost JA (2015) Acetylation of the RhoA GEF Net1A controls its subcellular localization and activity. J Cell Sci 128:913–922
Tao JJ, Chen HW, Ma B, Zhang WK, Chen SY, Zhang JS (2015) The role of ethylene in plants under salinity stress. Front Plant Sci 6:1–12
Torrigiani P, Serafini-Fracassini D, Biondi S, Bagni N (1986) Evidence for the subcellular localization of polyamines and their biosynthetic enzymes in plant cells. J Plant Physiol 124:23–29
Torrigiani P, Scaramagli S, Ziosi V, Mayer M, Biondi S (2005) Expression of an antisense Datura stramonium S-adenosylmethionine decarboxilase cDNA in tobbaco: changes in enzyme activity, putrescine-spermidine ratio, rhizogenic potential, and response to methyl jasmonate. J Plant Physiol 162:559–571
Truong K, Lee TD, Li B, Chen Y (2012) Sumoylation of SAE2 C terminus regulates SAE nuclear localization. J Biol Chem 287:42611–42619
Urano K, Yoshiba Y, Nanjo T, Igarashi Y, Seki M, Sekiguchi F, Yamaguchi-Shinozaki K, Shinozaki K (2003) Characterization of Arabidopsis genes involved in biosynthesis of polyamines in abiotic stress responses and developmental stages. Plant Cell Environ 26:1917–1926
Velikova V, Yordanov I, Edreva A (2000) Oxidative stress and some antioxidant system in acid rain treated bean plants: protective role of exogenous polyamines. Plant Sci 151:59–66
Waie B, Rajam MV (2003) Effect of increased polyamine biosynthesis on stress responses in transgenic tobacco by introduction of human S-adenosylmethionine gene. Plant Sci 164:727–734
Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334:1081–1086
Warton K, Foster NC, Gold WA, Stanley KK (2004) A novel gene family induced by acute inflammation in endothelial cells. Gene 342:85–89
Wen XP, Ban Y, Inoue H, Matsuda N, Kita M, Moriguchi T (2011) Antisense inhibition of a spermidine synthase gene highlights the role of polyamine for stress alleviation in pear shoots subjected to salinity and cadmium. Environ Exp Bot 72:157–166
Wi SJ, Kim WT, Park KY (2006) Overexpression of carnation S-adenosylmethionine decarboxylase gene generates a broadspectrum tolerance to abiotic stresses in transgenic tobacco plants. Plant Cell Rep 25:1111–1121
Wi SJ, Kim SJ, Kim WT, Park KY (2014) Constitutive S-adenosylmethionine decarboxylase gene expression increases drought tolerance through inhibition of reactive oxygen species accumulation in Arabidopsis. Planta 30:30
Woodgett JR, Gould KL, Hunter T (1986) Substrate specificity of protein kinase C. Eur J Biochem 161:177–184
Xue Y, Liu Z, Gao X, Jin C, Wen L, Yao X, Ren J (2010) GPS-SNO: Computational prediction of protein S-nitrosylation sites with a modified GPS algorithm. PLoS ONE 5:e11290
Yamanoha B, Cohen SS (1985) S-adenosylmethionine decarboxylase and spermidine synthase from Chinese cabbage. Plant Physiol 78:784–790
Yu Y, Jin C, Sun C, Wang J, Ye Y, Zhou W, Lu L, Lin X (2016) Inhibition of ethylene production by putrescine alleviates aluminium-induced root inhibition in wheat plants. Sci Rep 6:18888
Zheng L, Yao J, Gao F, Chen L, Zhang C, Lian L, Xie L, Wu Z, Xie L (2016) The subcellular localization and functional analysis of Fibrillarin2, a nucleolar protein in Nicotiana benthamiana. Biomed Res Int 2016:2831287
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgements
Open access funding provided by Szent István University (SZIE). This work was supported by grants from the Hungarian Scientific Research Fund (OTKA 101195) and Higher Education Institutional Excellence Program (20430-3/2018/FEKUTSTRAT) awarded by the Ministry of Human Capacities within the framework of plant breeding and plant protection researches of Szent István University, 2017-1.3.1-VKE-2017-0030 and the EFOP-3.6.3-VEKOP-16-2017-00008 project (co-financed by the European Union and the European Social Fund).
Author information
Authors and Affiliations
Contributions
The authors performed the following tasks: LK—designed the research, performed research, analyzed data, wrote the paper; ÁM—performed research, analyzed data; ASZ—performed research, analyzed data; SF—performed research; FS—performed research; KP—designed the research; EK—designed the research, analyzed data, wrote the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kovács, L., Mendel, Á., Szentgyörgyi, A. et al. Comparative analysis of overexpressed Fragaria vesca S-adenosyl-l-methionine synthase (FvSAMS) and decarboxylase (FvSAMDC) during salt stress in transgenic Nicotiana benthamiana. Plant Growth Regul 91, 53–73 (2020). https://doi.org/10.1007/s10725-020-00587-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-020-00587-3