Abstract
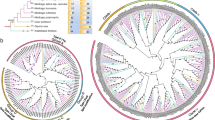
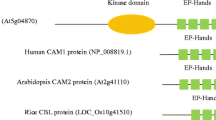
Calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs) form the CBL-CIPK complexes, perceiving calcium signals and relaying the signals to downstream responses in plants. To further understand the CBL-CIPK signaling system, here we focused on the evolutionary analysis of CIPKs. We re-evaluated eight plant genomes and identified 146 CIPKs, providing several new CIPKs in rice and poplar. A phylogenetic tree was constructed, showing that these 146 CIPKs are grouped into intron-rich and intron-less clades. Furthermore, all the CIPKs from the non-angiosperm species were found in intron-rich clade. We identified 30 conserved protein motifs among these 146 CIPKs. Analysis of gene duplication showed that the expansion of CIPKs in both clades is partly contributed by segmental duplications, however, tandem duplicates were found only in intron-less clade. Ka/Ks ratios showed that CIPK genes have experienced purifying selective pressure. Additionally, clustering of gene expression revealed that some CIPK genes in two clades share similar expression patterns under abiotic stresses and four CIPKs in intron-less clade form a distinct cluster (i.e., different expression patterns), suggesting the complexity of CIPK gene expression under abiotic stresses. Taken together, our results provided some new insights into the evolution of CIPKs and the hint that the expansion of CIPKs in intron-less clade is adaptive to environmental stresses.




Similar content being viewed by others
Abbreviations
- CBLs:
-
Calcineurin B-like proteins
- CIPKs:
-
CBL-interacting protein kinases
- HMM:
-
Hidden Markov model
- CAMs:
-
Calmodulins
- CDPKs:
-
Calcium-dependent protein kinases
- SOS:
-
Salt overly sensitive
References
Albrecht V, Ritz O, Linder S, Harter K, Kudla J (2001) The NAF domain defines a novel protein–protein interaction module conserved in Ca2+-regulated kinases. EMBO J 20:1051–1063
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intel Syst Mol Biol 2:28–36
Batistic O, Kudla J (2004) Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219:915–924
Chen X, Gu Z, Xin D, Hao L, Liu C, Huang J, Ma B, Zhang H (2011) Identification and characterization of putative CIPK genes in maize. J Genet Genomics 38:77–87
Demuth JP, Hahn MW (2009) The life and death of gene families. BioEssays 31:29–39
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14:755–763
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183:557–564
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Halfter U, Ishitani M, Zhu JK (2000) The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc Natl Acad Sci USA 97:3735–3740
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
Harmon AC, Gribskov M, Harper JF (2000) CDPKs—a kinase for every Ca2+ signal? Trends Plant Sci 5:154–159
Herrero J, Valencia A, Dopazo J (2001) A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17:126–136
Hrabak EM, Chan CW, Gribskov M, Harper JF, Choi JH, Halford N, Kudla J, Luan S, Nimmo HG, Sussman MR, Thomas M, Walker-Simmons K, Zhu JK, Harmon AC (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol 132:666–680
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, Finn RD, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Laugraud A, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, Mistry J, Mitchell A, Mulder N, Natale D, Orengo C, Quinn AF, Selengut JD, Sigrist CJ, Thimma M, Thomas PD, Valentin F, Wilson D, Wu CH, Yeats C (2009) InterPro: the integrative protein signature database. Nucleic Acids Res 37:D211–D215
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50:347–363
Kolukisaoglu U, Weinl S, Blazevic D, Batistic O, Kudla J (2004) Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol 134:43–58
Kudla J, Xu Q, Harter K, Gruissem W, Luan S (1999) Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc Natl Acad Sci USA 96:4718–4723
Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W (2002) Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14(Suppl):S389–S400
Ohta M, Guo Y, Halfter U, Zhu JK (2003) A novel domain in the protein kinase SOS2 mediates interaction with the protein phosphatase 2C ABI2. Proc Natl Acad Sci USA 100:11771–11776
Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD (2012) The Pfam protein families database. Nucleic Acids Res 40:D290–D301
Roy SW, Penny D (2007) Patterns of intron loss and gain in plants: intron loss-dominated evolution and genome-wide comparison of O. sativa and A. thaliana. Mol Biol Evol 24:171–181
Sanders D, Pelloux J, Brownlee C, Harper JF (2002) Calcium at the crossroads of signaling. Plant Cell 14(Suppl):S401–S417
Schumann N, Navarro-Quezada A, Ullrich K, Kuhl C, Quint M (2011) Molecular evolution and selection patterns of plant F-box proteins with C-terminal kelch repeats. Plant Physiol 155:835–850
Shi J, Kim KN, Ritz O, Albrecht V, Gupta R, Harter K, Luan S, Kudla J (1999) Novel protein kinases associated with calcineurin B-like calcium sensors in Arabidopsis. Plant Cell 11:2393–2405
Snedden WA, Fromm H (2001) Calmodulin as a versatile calcium signal transducer in plants. New Phytol 151:35–66
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wang D, Zhang Y, Zhang Z, Zhu J, Yu J (2010) KaKs_Calculator 2.0: a toolkit incorporating gamma-series methods and sliding window strategies. Genomics Proteomics Bioinform 8:77–80
Weinl S, Kudla J (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184:517–528
Xiang Y, Huang Y, Xiong L (2007) Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol 144:1416–1428
Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125:1347–1360
Yang X, Kalluri UC, Jawdy S, Gunter LE, Yin T, Tschaplinski TJ, Weston DJ, Ranjan P, Tuskan GA (2008) The F-box gene family is expanded in herbaceous annual plants relative to woody perennial plants. Plant Physiol 148:1189–1200
Ye CY, Li T, Tuskan GA, Tschaplinski TJ, Yang X (2011) Comparative analysis of GT14/GT14-like gene family in Arabidopsis, Oryza, Populus, Sorghum and Vitis. Plant Sci 181:688–695
Ye CY, Li T, Yin H, Weston DJ, Tuskan GA, Tschaplinski TJ, Yang X (2012) Evolutionary analyses of non-family genes in plants. Plant J. doi:10.1111/tpj.12073
Yu Y, Xia X, Yin W, Zhang H (2007) Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul 52:101–110
Acknowledgments
This research was supported by grants from the Ministry of Science and Technology of China (2011BAD38B01, 2009CB119101), the National Natural Science Foundation of China (31070597, 31270656, and 31100492), Joint Programs of the Scientific Research and Graduate Training from BMEC (Stress Resistance Mechanism of Poplar), and Beijing Forestry University Technology Innovation Program (BLYJ200904).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ye, CY., Xia, X. & Yin, W. Evolutionary analysis of CBL-interacting protein kinase gene family in plants. Plant Growth Regul 71, 49–56 (2013). https://doi.org/10.1007/s10725-013-9808-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10725-013-9808-5