Abstract
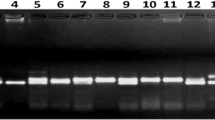
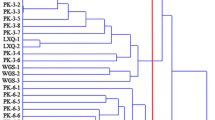
Okra [Abelmoschus esculentus (L.) Monech.] is an annual herb of the family Malvaceae and has tremendous nutritional and industrial potential. However, the unknown genome size, variation in ploidy level makes it more challenging for okra breeders to enhance their genetic potential. In this context, the genetic analysis of allelic polymorphism and population structure analysis of okra germplasm is pivotal for its genetic improvement. In the present investigation, a total of 96 okra genotypes were undertaken for morpho-genetic diversity analyses using 13 morphological parameters and 65 SSR markers. Out of 65 SSRs, 50 primers were polymorphic and the mean polymorphic information content (PIC) value was 0.38. The polymorphic loci AVRDC OKRA 64 revealed a maximum PIC value of 0.71 and the number of alleles per locus varied from 2 to 7, with an average of 3.34 alleles per locus. The neighbour-joining (N-J) tree grouped the genotypes into three distinct clusters (I-III), of which Cluster I comprised of cultivated species, A. esculentus and A. caillei, while, Cluster II and III consisted of other related wild species. Furthermore, the population structure analysis distinguished the okra genotypes into two distinct genetic groups comprising cultivated (A. esculentus) and wild-type okra. The variability analysis based on 13 agro-morphological traits revealed that the genotype DOV-92 exhibited the highest per se performance for total fruit yield, followed by Pusa Bhindi-5. The PCA (Principal Component Analysis) analysis revealed that the first three components account for more than 50% of the total genetic variation. The present investigation deciphered a high morphological and genetic diversity in cultivated and wild okra germplasm. Thus, it could serve as important genetic resources for okra improvement programmes.







Similar content being viewed by others
References
Adams CF (1975) Nutritive value of American foods in common units. Agricultural Research Service, US Department of Agriculture
Aladele SE, Ariyo OJ, de La Pena R (2008) Genetic relationships among West African okra (Abelmoschus caillei) and Asian genotypes (Abelmoschus esculentus) using RAPD. Afr J Biotechnol 7:1426–1431
Andries JA, Jones JE, Sloane LW, Marshall JG (1969) Effects of okra leaf shape on boll rot, yield, and other important characters of upland cotton Gossypium hirsutum L 1. Crop Sci 9(6):705–710
Ariyo OJ (1990) Effectiveness and relative discriminatory abilities of techniques measuring genotype × environment interaction and stability in okra [Abelmoschus esculentus (L) Moench]. Euphytica 47(2):99–105
Bhat KV, Bisht, IS, Mahajan RK, Rana RS (1996) Analysis of the genetic relationship among Abelmoschus species using isozyme and RAPD markers. In 2nd International Crop Science Congress, pp 17–24
Bisht IS, Mahajan RK, Rana RS (1995) Genetic diversity in South Asian okra (Abelmoschus esculentus) germplasm collection. Ann Appl Biol 126:539–550
Bisht IS, Bhat KV (2006) Okra (Abelmoschus spp.). In: Singh RJ (ed) Genetic Resources, Chromosome Engineering, and Crop Improvement: Vegetable crops, 3rd Vol. CRC press, pp 148–162
Cheng S, Melkonian M, Smith SA, Brockington S, Archibald JM, Delaux PM, Li FW, Melkonian B, Mavrodiev EV, Sun W, Fu Y (2018) 10KP: A phylodiverse genome sequencing plan. Gigascience 7(3):giy013
Cornille A, Feurtey A, Ge´lin U, Ropars J, Misvanderbrugge K, Gladieux P, Giraud T (2015) Anthropogenic and natural drivers of gene flow in a temperate wild fruit tree: a basis for conservation and breeding programs in apples. Evol Appl 8(4):373–384
Das A, Yadav R, Choudhary H, Singh S, Khade Y, Chandel R (2020) Determining genetic combining ability, heterotic potential and gene action for yield contributing traits and Yellow Vein Mosaic Virus (YVMV) resistance in Okra [Abelmoschus esculentus (L.) Monech.]. Plant Genet Resour: Characterisation Util 1:14. Doi: https://doi.org/10.1017/S1479262120000337
Das A (2019) Genetic Diversity based on Morphological and Molecular Markers andHeterosis Studies in Okra [Abelmoschus esculentus L.(Moench)]. M. Sc. Dissertation, Division of Vegetable Science, ICAR-Indian Agricultural Research Institute, New Delhi-12
Datta PC, Naug A (1968) A few strains of Abelmoschus esculentus (L.) Moench their karyological in relation to phylogeny and organ development. Beitr Biol Pflanz (BIOSIS) 45:113–126
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Düzyaman E (1997) Okra: botany and horticulture. Hortic Rev 21:41–72
Düzyaman E (2005) Phenotypic diversity within a collection of distinct okra (Abelmoschus esculentus) cultivars derived from Turkish landraces. Genet Resour Crop Evol 52:1019–1030
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Evanno G, Regnaut S, Goude J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. PMID: 15969739. Doi: https://doi.org/10.1111/j.1365-294x.2005.02553.x
FAOSTAT (2014) Available online at: http://faostat.fao.org/ (Accessed February 8, 2017)
Ford CE (1938) A contribution to a cytogenetical survey of the Malvaceae. Genetica 20:431–452
Fougat RS, Purohit AR, Kumar S, Parekh MJ, Kumar M (2015) SSR based genetic diversity in Abelmoschus species. Indian J Agr Sci 85:1223–1228
Fufa N (2019) Propagation methods of okra (Abelmoschus esculentus L.) and its application used in vitro plant regeneration”. Actasciagric 3:125–130
Gabriel KR (1971) The biplot graphic display of matrices with application to principal component analysis. Biometrika 58(3):453–467. https://doi.org/10.1093/biomet/58.3.453
Gangopadhyay KK, Singh A, Bag MK, Ranjan P, Prasad TV, Roy A, Dutta M (2017) Diversity analysis and evaluation of wild Abelmoschus species for agro-morphological traits and major biotic stresses under the north western agro-climatic condition of India. Genet Resour Crop Evol 64(4):775–790
Gulsen O, Karagul S, Abak K (2007) Diversity and relationships among Turkish okra germplasm by SRAP and phenotypic marker polymorphism. Biologia 62(1):41–45
Hazra P, Basu D (2000) Genetic variability, correlation and path analysis in okra. Ann Agric Sci 21:452–453
Huang J, Liu S, Guo X, Zhao YK, Li M, Zhang C, Zhu J, Ye J (2017) Genetic diversity and evolutionary analysis of okra [Abelmoschus esculentus (L.) Moench] germplasm resources based on ISSR markers. Genet Mol Res 1:1–8
Jaiprakashnarayan RP, Mulge R (2004) Correlation and path analysis in okra [Abelmoschus esculentus (L.) Moench]. Indian J Hortic 61:232–235
Joshi AB, Hardas MW (1956) Alloploid Nature of Okra [Abelmoschus esculentus (L.) Monech.]. Nature. 178(4543):1190
Joshi AB, Hardas MW (1953) Chromosome number of Abelmoschus tuberculatus Pal et Singh—a species related to the cultivated bhindi. Curr Sci 22(12):384–385
Kaur K, Pathak M, Kaur S, Pathak D, Chawla N (2013) Assessment of morphological and molecular diversity among okra [Abelmoschus esculentus (L.) Moench.] germplasm. Afr J Biotechnol 12:27–34
Kumar S, Parekh MJ, Fougat RS, Patel SK, Patel CB, Kumar M, Patel BR (2017) Assessment of genetic diversity among okra genotypes using SSR markers. J Plant Biochem Biot 26:172–178
Kumar C, Singh SK, Singh R, Pramanick KK, Verma MK, Srivastav M, Tiwari G, Choudhury DR (2019) Genetic diversity and population structure analysis of wild Malus genotypes including the crabapples (M. baccata (L.) Borkh. & M. sikkimensis (Wenzig) Koehne ex C. Schneider) collected from the Indian Himalayan region using microsatellite markers. Genet Resour Crop Evol 66(6):1311–1326
Kyriakopoulou OG, Arens P, Pelgrom KT, Karapanos I, Bebeli P, Passam HC (2014) Genetic and morphological diversity of okra [Abelmoschus esculentus [L.] Moench.] genotypes and their possible relationships, with particular reference to Greek landraces. Sci Hortic 171:58–70
Lever J, Krzywinski M, Altman N (2017) Principal component analysis. Nat Methods 14:641–642. https://doi.org/10.1038/nmeth.4346
Li FP, Lee YS, Kwon SW, Li G, Park YJ (2014) Analysis of genetic diversity and trait correlations among Korean landrace rice (Oryza sativa L.). Genet Mol Res 13(3):6316–6331
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
López-Cortés XA, Matamala F, Maldonado C, Mora-Poblete F, Scapim CA (2020) A deep learning approach to population structure inference in Inbred Lines of Maize. Front. Genet. 11:543459. Makumbi D, Betran JF, Banziger M, Ribaut JM (2011) Combining ability, heterosis and genetic diversity in tropical maize (Zea mays L.) under stress and non-stress conditions. Euphytica 180:143–162
Mishra GP, Singh B, Seth T, Singh AK, Halder J, Krishnan N, Tiwari SK, Singh PM (2017) Biotechnological advancements and begomovirus management in okra (Abelmoschus esculentus L.): Status and perspectives. Front Plant Sci 8:360
Nadkarni KM (1927) Indian Meteria Medica. Nadkarni and Co.
Omasheva MY, Flachowsky H, Ryabushkina NA, Pozharskiy AS, Galiakparov NN, Hanke MV (2017) To what extent do wild apples in Kazakhstan retain their genetic integrity? Tree Genet Genomes 13(3):52
One Thousand Plant Transcriptomes Initiative (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574(7780):679
Ostrowski MF, David J, Santoni S, McKhann H, Reboud X, Le Corre V, Camilleri C, Brunel D, Bouchez D, Faure B, Bataillon T (2006) Evidence for a large-scale population structure among accessions of Arabidopsis thaliana: possible causes and consequences for the distribution of linkage disequilibrium. Mol Ecol 15(6):1507–1517
Park YH, West MA, St. Clair DA (2004) Evaluation of AFLPs for germplasm fingerprinting and assessment of genetic diversity in cultivars of tomato (Lycopersicon esculentum L.). Genome 47(3):510–518
Peakall RO, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Poehlman JM, Sleper DA (1995) Breeding of Field Crops. 4th edn. Iowa State University Press, Ames, IA
Prakash K, Pitchaimuthu M, Ravishankar KV (2011) Research Article Assessment of genetic relatedness among okra genotypes [Abelmoschus esculentus (L.) Moench] using RAPD markers. Electron J Plant Breed 2(1):80–86
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959 (PMID: 10835412)
R Studio Team (2020) RStudio: Integrated Development for R. RStudio, PBC, Boston, MA URL. http://www.rstudio.com/
Ramya P, Bhat KV (2012) Analysis of phylogenetic relationships in Abelmoschus species (Malvaceae) using ribosomal and chloroplast intergenic spacers. Indian J Genet Plant Breed 72(4):445–453
Ravishankar KV, Muthaiah G, Mottaiyan P, Gundale SK (2018) Identification of novel microsatellite markers in okra [Abelmoschus esculentus (L.) Moench] through next-generation sequencing and their utilization in analysis of genetic relatedness studies and cross-species transferability. J Genet 97(1):39–47
Rohlf FJ (1998) NTSYSpc: numerical taxonomy and multivariate analysis system. Version 2.02. Exeter Software, Setauket
Salameh MN (2014) Genetic Diversity of Okra (Abelmoschus esculentus L.) landraces from different agro-ecological regions revealed by AFLP analysis. Am Eurasian J Agric Environ Sci 14:155–160
Schafleitner R, Kumar S, Lin CY, Hegde SG, Ebert A (2013) The okra (Abelmoschus esculentus) transcriptome as a source for gene sequence information and molecular markers for diversity analysis. Gene 517:27–36
Sethy NK, Shokeen B, Edwards KJ, Bhatia S (2006) Development of microsatellite markers and analysis of intraspecific genetic variability in chickpea (Cicer arietinum L.). Theor Appl Genet 112(8):1416–1428
Shetty AA, Singh JP, Singh D (2013) Resistance to yellow vein mosaic virus in okra: A review. Biol Agric Hortic 29:159–164
Siemonsma JS (1982) West african okra—Morphological and cytogenetical indications for the existence of a natural amphidiploid of Abelmoschus esculentus (L.) Moench and A. manihot (L.) Medikus. Euphytica 31:241–252
Singh B, Aakansha G (2014) Analysis of genetic diversity in okra [Abelmoschus esculentus (L.) Moench] genotypes using RAPD markers. Vegetos 27:266–271
Singh K, Pandey UB (1993) Export of vegetables-status and strategies. Veg Sci 20:93–103
Singh B, Chaubey T, Upadhyay DK, Jha AASTIK, Pandey SD, Sanwal SK (2015) Varietal characterization of okra (Abelmoschus esculentus) based on morphological descriptions. Indian J Agric Sci 85(9):1192–1200
Singh BD, Singh AK (2015) Marker Assisted Plant Breeding: Principles and Practices. Springer, pp 217–255
Verma H, Borah JL, Sarma RN (2019) Variability assessment for root and drought tolerance traits and genetic diversity analysis of rice germplasm using SSR markers. Sci Rep 9:16513. https://doi.org/10.1038/s41598-019-52884-1
Yadav RK, Badiger M, Lata S (2019) Studies of genetic variability and association of yield traits and YVMV disease in Abelmoschus species. Indian J Agric Sci 89(4):688–694
Yıldız M, Koçak M, Baloch FS (2015) Genetic bottlenecks in Turkish okra germplasm and utility of iPBSretrotransposon markers for genetic diversity assessment. Genet Mol Res 14(3):10588–10602
Yıldız M, Ekbiç E, Düzyaman E, Serçe S, Abak K (2016) Genetic and phenotypic variation of Turkish Okra (Abelmoschus esculentus L. Moench) accessions and their possible relationship with American, Indian and African germplasms. J Plant Biochem Biot 25(3):234–244
Yonas M, Garedew W, Debela A (2014) Multivariate analysis among okra [Abelmoschus esculentus (l.) moench] collection in South Western Ethiopia. J Plant Sci 9(2):43
Yuan CY, Zhang C, Wang P, Hu S, Chang HP, Xiao WJ, Guo XH (2014) Genetic diversity analysis of okra (Abelmoschus esculentus L.) by inter-simple sequence repeat (ISSR) markers. Genet Mol Res 13:3165–3175
Acknowledgements
The authors are highly thankful to the Division of Vegetable Science and PG School, ICAR-IARI, New Delhi, to provide all sorts of laboratory facilities and financial support. First Author also wants to extend his acknowledgement to the Indian Council of Agriculture Research (ICAR) for awarding ICAR-PG Fellowship-2017 during the tenure of the Post-graduate research program.
Funding
There is no specific financial assistance provided for this work.
Author information
Authors and Affiliations
Contributions
Anjan Das: Investigation, Data Curation, Formal analysis, Writing—Original Draft; Ramesh Kumar Yadav: Conceptualization, Resources, Supervision, Writing—Review & Editing; Harshwardhan Choudhary: Writing—Review & Editing, Methodology; Suman Lata: Formal analysis, Visualization, Writing—Review & Editing; Saurabh Singh: Formal analysis, Visualization; Writing—Review & Editing; Chavlesh Kumar: Formal analysis; Shilpi Kumari: Formal analysis, Data Curation; Boopalakrishnan G: Data Curation; Rakesh Bhardwaj: Data Curation, AkshayTalukdar: Formal analysis.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Human and animal rights
No animal or any human participants were used.
Informed consent
All the authors were informed and they have given the consent for the publication in this journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Das, A., Yadav, R.K., Choudhary, H. et al. Population structure, gene flow and genetic diversity analyses based on agro-morphological traits and microsatellite markers within cultivated and wild germplasms of okra [Abelmoschus esculentus (L.) Moench.]. Genet Resour Crop Evol 69, 771–791 (2022). https://doi.org/10.1007/s10722-021-01263-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-021-01263-9