Abstract
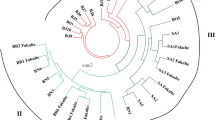
Prunus sibirica (Siberian apricot) has ecological, economic, and social benefits that makes it an important fruit-bearing tree in China. However, current fruit yields are low and unstable, thus restricting development of Prunus sibirica industry. To provide a scientific basis for successful breeding research aimed at developing superior cultivars, 98 screened SSR markers were first used in our study, developed by restriction-site associated DNA sequencing, to assess the genetic diversity of 66 Prunus sibirica accessions collected from four populations. The average number of alleles per locus (9.910) and the number of effective alleles (5.445) showed high polymorphism in the entire population, and the polymorphism information content (0.675) indicated that these markers were highly polymorphic. There was gene flow among the Prunus sibirica accessions, however the genetic differentiation coefficient showed 15.4% gene frequency differentiation among the provenances. Meanwhile, extensive linkage disequilibrium (D′ > 0.5, P < 0.01) was found, however the overall level was low (r2 < 0.5, p < 0.01). Additionally, the 66 accessions clustered into four groups, and these groups were extremely significantly correlated with the provenances classification. The clustering results showed that geographical distribution and genetic diversity changed from high to low as the geographic separation between provenances increased. Furthermore, population structure analysis supported these findings as genetic structure and provenances were extremely significantly correlated (p < 0.01). Additionally, the relationships between the geographical and genetic distances of the provenances and of the individuals were significantly correlated, indicating that geographical isolation importantly influenced Prunus sibirica evolution. The geographic and genetic effects underlying the superior accessions which we selected provide a reference for future molecular marker assisted breeding of Prunus sibirica.



Similar content being viewed by others
References
Barchi L, Lanteri S, Portis E, Acquadro A, Valè G, Toppino L, Rotino GL (2011) Identification of SNP and SSR markers in eggplant using RAD tag sequencing. BMC Genomics 12:304
Bolger A, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30:2114–2120
Bradbury PJ, Zhang ZW, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Cardon LR, Bell JI (2001) Association study designs for complex diseases. Nat Rev Genet 2:91–98
Dong SB, Liu YL, Niu J, Ning Y, Lin SZ, Zhang ZX (2014) De novo transcriptome analysis of the Siberian apricot (Prunus sibirica L.) and search for potential SSR markers by 454 pyrosequencing. Gene 544(2):220–227
Du QZ (2014) Dissection of allelic variation underlying important traits in Populus tomentosa Carr. by using joint linkage and linkage disequilibrium mapping. Dissertation/Master’s Thesis. Beijing Forestry University
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Filippi CV, Merino GA, Montecchia JF, Aguirre NC, Rivarola M, Naamati G, Fass MI, Álvarez D, Rienzo JD, Heinz RA, Moreira BC, Lia VV, Paniego NB (2020) Genetic diversity, population structure and linkage disequilibrium assessment among international sunflower breeding collections. Genes 11(3):283
Flint-Garcia SA, Thornsberry JM, Buckler ES (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 06:13
Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39:1151–1155
Li M, Zhao Z, Miao XJ, Zhou JJ (2014) Genetic diversity and population structure of Siberian apricot (Prunus sibirica L.) in China. Int J Mol Sci 15:377–400
Li M, Zheng PG, Ni BY, Hu X, Miao XJ, Zhao Z (2018) Genetic diversity analysis of apricot cultivars grown in China based on SSR markers. Eur J Hortic Sci 83:18–27
Liu MP (2011) Genetic diversity of Armeniaca cathayana based on SSR and ISSR analysis. Dissertation/Master’s thesis. Beijing Forestry University
Liu KJ, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21(9):2128–2129
Liu HB, Liu J, Wang Z, Ma LY, Wang SQ, Lin XG, Wu RL, Pang XM (2013) Development and characterization of microsatellite markers in Prunus sibirica (Rosaceae). Appl Plant Sci 1(3):253–255
Malosetti M, van der Linden CG, Vosman B, van Eeuwijk FA (2007) A mixed-model approach to association mapping using pedigree information with an illustration of resistance to Phytophthora infestans in potato. Genetics 175:879–889
Mantel N (1967) Ranking procedures for arbitrarily restricted observation. Biometrics 23:65–78
Miller MP, Knaus BJ, Mullins TD, Haig SM (2013) SSR pipeline: A bioinformatic infrastructure for identifying microsatellites from paired-end Illumina high-throughput DNA sequencing data. J Hered 104:881–885
Neale DB, Savolainen O (2004) Association genetics of complex traits in conifers. Trends Plant Sci 09:325–330
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Neilsen R (2005) Molecular signatures of natural selection. Ann Rev Gen 39:197–218
Niu SZ, Song QF, Koiwa H, Qiao DH, Zhao DG, Chen ZW, Liu X, Wen XP (2019) Genetic diversity, linkage disequilibrium, and population structure analysis of the tea plant (Camellia sinensis) from an origin center, Guizhou plateau, using genome-wide SNPs developed by genotyping-by-sequencing. BMC Plant Biol 19(1):328
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rubio-Moraga A, Cande-Perez D, Lucas-Borja ME, Tiscar PA, Viñegla B, Linares JC, Gómez-Gómez L, Ahrazem O (2012) Genetic diversity of Pinus nigra Arn. Populations in Southern Spain and Northern Morocco revealed by inter-simple sequence repeat profiles. Int J Mol Sci 13(5):5465–5658
Sanchez M, Ingrouille MJ, Cowan RS, Hamilton MA, Fay MF (2014) Spatial structure and genetic diversity of natural populations of the Caribbean pine, Pinus caribaea var bahamensis (PInaceae), in the Bahaman archipelago. Bot J LInnean Socirty 174(3):359–383
Smith JSC, Chine CL, Shu H (1997) An evaluation of the utility of SSR loci as molecular markers in maize (Zea mays L.): comparisons with data from RFLPs and pedigree. Theor Appl Genet 95:163–173
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Thistlethwaite FR, El-Dien OG, Ratcliffe B, Klápště J, Porth I, Chen C, Stoehr MU, Ingvarsson PK, El-Kassaby YA (2020) Linkage disequilibrium vs. pedigree: genomic selection prediction accuracy in conifer species. PLoS ONE 15(6):e0232201
Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2014) Primer3: new capabilities and interfaces. Nucl Acids Res 40(15):e115
Wang MX (2001) Forest tree genetics and breeding. China Forestry Publishing House, Beijing
Wang YX (2008) A molecular genetic linkage map for hybrid population of Populus adenopoda × P. alba and a comparative map in Populus. Dissertation/Doctor’s thesis. Nanjing Forestry University
Wang CH, Tian YK, Zhao J (2005) General application analysis of SSRs derived from apple (Malus pumila) on other species in Rosaceae. Acta Hortic Sin 32:500–502
Wang J, Wei AZ, Yang TX (2011) The analysis and comprehensive valuation on manyquantitative characters of different apricots. Northern Hortic 12:5–9
Wang Z, Liu HB, Liu J, Li YY, Wu RL, Pang XM (2014) Mining new microsatellite markers for Siberian apricot (Prunus sibirica L.) from SSR-enriched genomic library. Sci Hortic 16:65–69
Yu HP (2013) Development of simple sequence repeat (SSR) markers from Paeonia ostia and its unilization in genetic relationshio analysis. Dissertation/Master’s thesis. Beijing Forestry University
Zhang HX, Weng JF, Zhang XC, Liu CL, Yong HJ, Hao ZF, Li XH (2014) Genome-wide association analysis of kernel row number in maize. Acta Agron Sin 40:1–6
Funding
This work was supported by National Key Research and Development Program of China (SQ2019YFD100071).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Chen, J., Dong, S., Zhang, X. et al. Genetic diversity of Prunus sibirica L. superior accessions based on the SSR markers developed using restriction-site associated DNA sequencing. Genet Resour Crop Evol 68, 615–628 (2021). https://doi.org/10.1007/s10722-020-01011-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-020-01011-5