Abstract
Bananas (Musa spp.), including dessert and cooking types, are of major importance in the tropics. Due to extremely high levels of sterility, the diversity of cultivated bananas is fixed over long periods of time to the existing genotypes. This pattern puts banana-based agrosystems at risk. Therefore, assessing the extent of wild and cultivated banana diversity, conserving it and making it available for further use is a priority. We report here the collection of new wild and cultivated banana germplasm in the Autonomous Region of Bougainville, Papua New Guinea. In total, 61 accessions were collected and their names and uses were recorded when possible. Classification was also provided based on the observations made in the field. Three wild specimens were collected. Among the 58 cultivated accessions, we noted that eight were used as ornamental plants, seven were edible varieties of the Fe’i type and two were natural tetraploids from the Musa section. The ploidy was then checked by flow cytometry and the accessions were genotyped with a set of 19 SSR markers. The genotyping results were merged to the dataset from Christelová et al. (Biodivers Conserv 26:801–824, 2017). This joint analysis helped refine or confirm the classification of the collected accessions. It also allowed to identify 10 private alleles and 35 genotypes or Genotype Groups that were not present in the wider dataset. Finally, it shed light on the diversification processes at work in the region, such as the capture of mutations by farmers and the likely occurrence of geneflow within the cultivated genepool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bananas (Musa spp.), including dessert and cooking types such as Plantain, are of major importance in the tropics for both subsistence and food security (FAO 2014). Originating from the South-East Asia/West Oceania region, this crop has a complex domestication history (De Langhe et al. 2009; Perrier et al. 2011). The name “banana” corresponds to different species of the Musa genus, and to their hybrids. The genus Musa is divided into two sections corresponding to distinct phylogenetic clades, Musa (2n = 22) and Callimusa (2n = 20 or 18) (Häkkinen 2013; Janssens et al. 2016). The edible bananas from the section Musa are composed of either genome A (Musa acuminata Colla), or A in combination with B (M. balbisiana Colla) or S (M. schizocarpa N.W. Simmonds). Edible bananas from the Musa section are diploid, triploid or more rarely tetraploid, the most popular cultivars being triploid from Groups such as Cavendish (AAA) and Plantain (AAB). Edible bananas from the section Callimusa, called Fe’i bananas, are associated with T genome (M. textilis Née), but have been far less studied and their origin remains obscure. Recent results indicate that the Fe’i bananas arose from complex domestication schemes and comprise accessions with different ploidy (Christelová et al. 2017; www.crop-diversity.org/mgis/). As edible bananas bear seedless fruits, they are propagated vegetatively. This mode of propagation and extremely low level of fertility make cultivated bananas particularly vulnerable to diseases and abiotic stress and the occurrence of new, potentially better adapted genotypes is limited to rare mutation events. Therefore, the diversity of cultivated bananas was fixed over long periods of time to the existing genotypes. Even if the diverse genetic make-up of banana varieties grown worldwide exhibit a wide range of traits (Heslop-Harrison and Schwarzacher 2007), this pattern slows down cultivated bananas’ evolution and puts banana-based agrosystems at risk by hampering rapid adaptation of the crop to new threats.
Breeding programs in banana focus primarily on creating improved triploid or tetraploid varieties (Ortiz 2013; Tomekpe et al. 2004). In addition to multiple ploidy levels, the sterility associated with the production of seedless fruits is a challenge for breeders. However, the use of fully or partially fertile, parthenocarpic, edible diploids eases the process of creating progenies in a largely sterile crop (Tomekpe et al. 2004; Tenkouano et al. 2011). In this context, assessing the extent of wild and cultivated banana diversity, conserving it and making it available for further use is a priority.
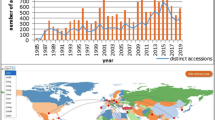
Papua New Guinea (PNG), including neighbouring islands, is a recognized centre of diversity and potentially a domestication centre for banana (Christelová et al. 2017; Lebot 1999; Sardos et al. 2016b). Four banana collecting missionsFootnote 1 were organized to mainland PNG and the Bismarck Archipelago in 1988–1989 which revealed high levels of diversity in the country. In total, 264 wild and cultivated accessions were collected, of which 86% appeared to be unique genotypes (Arnaud and Horry 1997). These accessions were sent to both the National Banana Germplasm Collection at Laloki, Port Moresby and to the Bioversity International Musa Germplasm Transit Centre (ITC)Footnote 2 in Belgium for conservation purposes. Currently, more than 25 years after the PNG missions, 230 of the accessions collected in PNG are still conserved in vitro in the ITC. Over the years, the PNG accessions have been valuable resources for breeders and researchers and have significantly improved our knowledge of banana (e.g. Ploetz et al. 1999; Geering et al. 2005; Raboin et al. 2005; Ball et al. 2006; Volkaert 2011; Till et al. 2010; Hřibová et al. 2011; Teo et al. 2011; Christelová et al. 2011, 2017; Valdez-Ojeda et al. 2014; Janssens et al. 2016; Sardos et al. 2016a, b).
Interestingly, out of the 177 cultivated accessions collected in PNG that are still conserved in the ITC, 100 are cultivated diploids with AA genome composition. These accessions are mainly used by local populations as cooking varieties and are therefore of great interest for the improvement of cooking triploids such as Plantains. Due to a civil conflict (1988–1998), the region which later became the Autonomous Region of Bougainville (AROB) was not visited by the expeditions in the 1980′s (Fig. 1).
Located in Near Oceania, the AROB is composed of two main islands, Bougainville and Buka, and of smaller surrounding islands. While it is an autonomous region of Papua New Guinea, it belongs geographically and ecologically to the Solomon Islands Archipelago. Agricultural systems in Bougainville and Buka are typical of the West Oceania region, where people practise a rain fed and shifting agriculture with fallow periods that can reach up to 15 years. The most important staple crop all over these islands is sweet potato (Ipomoea batatas (L.) Lam.) followed by banana, coconut (Cocos nucifera L.) and a range of root and tuber crops (Bourke et al. 2002).
Organized by NARI and Bioversity International, a banana collecting mission in the AROB took place from 19 October to 31 October 2016, and explored the islands of Bougainville, Buka and Sohano. This report presents the genetic diversity collected during the prospections, which targeted new wild and cultivated germplasm for conservation purposes. When possible, the prospections were coupled with the systematic ploidy measurement and SSR genotyping of the collected accessions. These two activities took place at the Musa Genotyping Centre (MGC) held in the Institute of Experimental Botany (Olomouc, Czech Republic). The SSR profiles of the collected accessions were added to the results obtained in the study published by Christelová et al. (2017) in order to better characterize the genetic diversity of the new accessions collected in this region. We discuss here the diversity of the genotypes collected in Bougainville and show that doubling plant prospection with “real-time” ploidy measurements and SSR genotyping adds substantial value to collecting missions.
Materials and methods
Collecting trip and documentation
The collecting trip was set up from 19 October to 31 October 2016, just before the rainy season. The team was composed of JS, JP, SJ, GSS and GR. Due to potential safety uncertainty, a local guide, Mr Zohn Bosco Miriona from Bougainville Experience Tours (BET), was contracted. Officers from Department of Primary Industries (DPI) offices in Buka, Kieta and Buin were also contacted to support the team locally.
The aim of this expedition was to seek new germplasm to enrich the national banana collection of NARI-Laloki and ultimately the ITC, in order to ensure the conservation and use of a wider diversity of Musa. The range of diversity encountered was documented (Sachter-Smith et al. 2017), but suckers and leaf samples were collected only when cultivated varieties were unknown to the team based on their morphology. Once suckers were collected, an accession code was provided and each accession was documented with local names, their meaning, origins, uses and GPS coordinates (Fig. 1). Potential classifications were also given based on the morphology of the plants. All collections, except from abandoned food gardens, were made with the authorization of the field’s owner, or a relative. The Fe’i types being quite uncommon, all Fe’i bananas encountered were collected along with a few samples of wild Callimusa previously described by Argent (1976) (M. bukensis Argent and M. maclayi F. Muell. subsp. maclayi var. erecta (N. W. Simmonds) Argent).
Sample processing
Once identified as potentially absent from the NARI collection, the varieties and wild specimens were given unique ID codes and became accessions. Suckers were collected for further ex-situ conservation and fresh leaf tissues, preferentially cigar leaves, were collected from all the accessions and conserved in an electric cooler that could be plugged to the car and in any guesthouse equipped with power. Two types of back-up samples were kept for DNA: one in DNAgard® Tissue (Biomatrica) http://www.biomatrica.com/media/dnagard_tissue/DNAgard-Tissue-preserves-plant-DNA.pdf and one silica dried. Fresh leaves were sent to the MGC-Olomouc through fast courier upon return of the team to Port Moresby. Back-ups were used for DNA extraction when necessary, i.e., when fresh leaves arrived at MGC that were too damaged.
Ploidy level estimation
Ploidy level of accessions for which fresh leaves were available was estimated by flow cytometry according to Doležel et al. (1997, 2007). About 30 mg of young leaf tissue was chopped with a razor blade in a Petri dish containing 500 µL Otto I solution (0.1 M citric acid, 0.5% v/v Tween 20). Crude homogenate was filtered through a 50 µm nylon mesh. Chicken red blood cell nuclei (CRBC), prepared according to Galbraith et al. (1998), were added to the suspension of banana nuclei as an internal standard. After 30 min incubation at room temperature, 1 mL Otto II solution (0.4 M Na2HPO4) (Otto 1990) supplemented with 2 µg/mL DAPI (4,6-diamidino-2-phenylindole). The samples were analysed using Sysmex-Partec CyFlow flow cytometer (Görlitz, Germany). The gain of the instrument was adjusted so that the peak of the CRBC nuclei was positioned approximately on channel 100 on a histogram of relative fluorescence intensity when using a 512-channel scale. The ploidy level of each banana accession was then determined by comparing peak positions of CRBC and Musa nuclei. The ratio between relative DAPI fluorescence intensity of CRBC nuclei and G1-phase nuclei of the accessions from the Musa section is ~ 0.5 for diploid plants and ~ 0.75 for triploid plants (Doležel et al. 1997; Christelová et al. 2017).
SSR genotyping
Leaf tissues were used for DNA extraction using NucleoSpin Plant II kit (Macherey–Nagel, Düren, Germany). Genotyping based on 19 SSR loci was performed according to Christelová et al. (2011). Briefly, all SSR loci were amplified from each DNA sample by PCR with locus-specific M13-tailed primer pair and fluorescently labelled M13 universal primer. PCR conditions were set as follows (in total volume of 20 μL): PCR reaction buffer (10 mM Tris-HCl, pH 8; 50 mM KCl; 0.1% Triton-X100; 1.5 mM MgCl2), 200 μM dNTPs, 1 U Taq polymerase (NEB), 8 pmol of the M13-tailed locus-specific forward primer, 10 pmol of the locus-specific reverse primer and 6 pmol of fluorescently labelled M13-universal primer. Four different fluorescent dyes (6-FAM, VIC, NED PET) for the M13 primer were used to allow for multiplexed fragment analysis of resulting PCR products. PCR was done in 35 cycles (94 °C/45 s, Ta/60 s, 72 °C/60 s) preceded by a denaturation step (94 °C/5 min) and followed by a final extension step (72 °C/5 min). Locus specific annealing temperature (Ta) followed Christelová et al. (2011). PCR products were purified by ethanol/sodium acetate precipitation and two independent PCR runs were done for each DNA sample.
Subsequently, purified PCR products were diluted 40-fold in Hi-Di formamide, mixed with internal standard GeneScan™– 500 LIZ size standard (Applied Biosystems, Foster City, CA, USA), denatured for 5 min at 95 °C and loaded onto capillary electrophoresis DNA analyzer (ABI 3730xl, Applied Biosystems, USA). Default module settings were used for electrophoretic separation and signal detection. Resulting raw data were processed by GeneMarker® v.1.75 (Softgenetics, State College, PA, USA) software to call alleles at individual SSR loci.
SSR data analysis
Due to the co-dominant nature of SSR markers and to the presence of several ploidy levels, the whole AROB dataset was coded as a binary presence (1) and absence (0) matrix and merged to a core dataset (CS) from Christelová et al. (2017). The CS used here is composed of 583 accessions with robust classification. Both the Musa Sect. (545 accessions) and the Callimusa Sect. (38 accessions) were represented.
The total number of alleles in the combined dataset was evaluated, the allelic patterns of the CS and the AROB datasets were compared and private alleles, i.e. alleles present in the AROB dataset but not in the CS, were identified. For the purpose of this paper, we then considered cultivated bananas from both sections separately.
The computer program DARwin 6 (Perrier and Jacquemoud-Collet 2006; Perrier et al. 2003) was then used to calculate dissimilarity values between pairs of accessions within a joint CS-AROB dataset using the Dice index. For this purpose we filtered the dataset on missing data and the marker mMaCIR164 which didn’t amplify 24% of the samples was excluded. Equally, 23 accessions exhibiting more than 20% of missing data were discarded from the set. All 23 accessions belonged to the Callimusa section (five cultivated Fe’i and 18 wild species). The CS was then pruned to avoid bias due to redundancies between identical genotypes or numerous closely related accessions. Notably, only six accessions of the Plantains Group were kept out of the 113 in the initial set. The pruned CS comprised 357 accessions from the section Musa including cultivated varieties and wild direct ancestors and 38 accessions belonging to the section Callimusa. We identified identical genotypes within the AROB dataset and between the AROB and the pruned CS datasets (dissimilarity = 0). Using the method proposed by Douhovnikoff and Dodd (2003) and a graph of the distribution of dissimilarity values with a step of 0.02 produced by DARwin 6 (S1 file), we also identified a dissimilarity threshold below which the genetic variation observed between accessions is considered to result either from genotyping errors or from the accumulation of mutations during the clonal propagation of a unique original genotype. This dissimilarity threshold was determined at 0.1 (S1 file) and allowed the identification of Genotype Groups (GGs) clustering around the AROB accessions and composed of nearly identical genotypes.
Due to high rates of missing data in the Callimusa section, we decided to only consider the edible bananas from the Musa section and their closely associated wild relatives. Using DARwin 6 and given the known history of cultivated bananas, we built a weighted Neighbor-Joining (NJ) tree under the topological constraint of a NJ tree built on the diploid accessions. The accessions for which ploidy was not measured were considered polyploids.
Results
Collection
In total and based on their morphology, the expedition collected 61 accessions probably not conserved in NARI, including three wild specimens belonging to the species M. bukensis (AROB002 and AROB008) and M. maclayi subsp. maclayi var. erecta (AROB013) (Table 1). Among the 58 cultivated accessions collected, four duplicate pairs that were not at the same developmental stage at the time of collection are suspected: AROB042 “Asi”/AROB045 “Glenda’s dwarf”, AROB043 “Sausage banana”/AROB061 “Sausage banana”, AROB029 “Kourai”/AROB031 “Kourai” and AROB051”Limot”/AROB052 “Poso-olohi”. In a few cases, names of the banana varieties were not known and the team named the accessions after the place of collection or after the owner of the variety.
Out of the cultivated banana varieties for which uses were documented, eight were used as ornamental among which a specimen belonging to the species M. ornata Roxb., 23 were preferentially used as cooking varieties, 12 were either cooked or used as dessert types, and nine were used preferentially as dessert varieties. We noted that among the cooking accessions collected, one was variegated, AROB055 “Tambra”. In total, nine accessions were named “wild banana” in local languages. Three of them were actually the wild accessions collected while the six others were found in cultivation, often near houses, and were classified as Fe’i bananas based on their morphology. However, among these Fe’i, two (AROB010 “Bia Kaura” and AROB030 “Korai 2”) were reported by farmers to bear a few seeds in the fruits and to have been collected from the wild. None of these accessions was flowering at the time of the prospection so it was not possible to strictly assign them to one of the local wild species.
The cultivated varieties collected were classified as AA (28), AAA (7), AAB (14) and two potential tetraploids (4x) were identified: AROB027 “Buka”, morphologically close to the Pisang Awak Group (ABB), and AROB056 “Kalmagol”, similar to the Silk Group (AAB). In addition to the two potential tetraploids, the team was not able to propose robust classifications for nine accessions. Four of them were red or reddish ornamental plants: AROB001 “Flower banana”, AROB006 “Nono 1”, AROB041 “Glenda’s Red” and AROB043 “Sausage Banana”. The other were edible types: AROB009 “Bukatawawe”, which was morphologically similar to ITC0605 “Japaraka n°2” (AA) but taller and therefore suspected to be AAA, AROB032 “Toitoi”, evoking an AAB but with dark green young fruits, AROB038 “Sinsiruai”, somewhat Maoli-Popoulu like and therefore suspected to be AAB, AROB057 “Sepik”, a very tall plant with a large bunch and a very unusual round and obtuse purple/yellow male bud and AROB061 “Sausage Banana” with fruits said to be red and sausage-like in appearance but were not observed.
Detailed results of the prospection are presented in Table 1 and pictures are available in Sachter-Smith et al. (2017).
Ploidy
Out of the 61 fresh leaf samples collected and sent to the MGC, 48 arrived in a good state to be used for ploidy measurement using flow cytometry. Overall, the results obtained were consistent with the classification determined based on the accessions’ morphology and allowed to refine a number of cases for which the team had doubts, notably confirming the tetraploid status of AROB027 “Buka” and AROB056 “Kalmagol”. Out of the 48 accessions, 34 were diploid. However, we noted that five of the seven Fe’i accessions collected and the accession AROB013 M. maclayi subsp. maclayi var. erecta exhibited peak ratios with internal standard slightly higher than expected for a regular diploid. Ploidy for these samples should ultimately be checked by chromosome counting but these results are not surprising for Callimusa accessions. It was shown that despite a lower number of chromosomes (x = 10), their genomes are larger than in the Musa section (x = 11) (Bartoš et al. 2005, Čížková et al. 2015).
SSR genotyping
Number of alleles
Out of the 61 samples sent to the MGC for genotyping, DNA extraction failed for AROB004 “Wiau”. We therefore obtained genotyping results for 60 accessions. Considering the Musa and Callimusa sections together, a total of 207 alleles was found in the AROB dataset while 353 alleles are present in the combined AROB—CS dataset. Out of the ten alleles only observed in the AROB accessions and not in the CS, seven were found within the Callimusa section and three in the Musa section (Fig. 2). In the Musa section, two of the new alleles were found in the triploid AAA AROB057 “Sepik” while the third one was found in the triploid AAA AROB017 “Banawa”. In the Callimusa section, new alleles were found in both the wild species and the Fe’i that were collected.
Distribution of all distinct alleles across 19 SSR loci for the Musa and Callimusa sections of the Core Set (CS) and of the Autonomous Region of Bougainville (AROB). Overlapping areas denote shared alleles between categories. Venn diagram constructed with the tool developed by Bardou et al. (2014)
Identical genotypes and Genotype Groups (GGs)
Out of 60 accessions genotyped, we identified 46 different genotypes or Genotype Groups (GGs), among which 35 were not present in the CS. Six pairs of strictly identical genotypes were identified within the AROB accessions collected (Tab. 2). In two cases, the genotyping confirmed what was suspected in the field (AROB042 “Asi”/AROB045 “Glenda’s Dwarf” and AROB051 “Limot”/AROB052 “Poso-olohi”) but in four cases it highlighted similarities that were not detected at collection (AROB018 “Tomea”/AROB033 “Papua”, AROB021 “Abau”/AROB028 “Popondetta”, AROB023 “Morou”/AROB055 “Tambra” and AROB059 “Goum”/AROB061 “Sausage Banana”). In addition, we identified two AROB genotypes that were strictly identical to ITC accessions: AROB009 “Bukatawawe” classified as AAA has the same genotype as ITC0372 “Hungtu” (AAA) while AROB028 “Popondeta” and AROB021 “Abau”, both classified as AA, have genotypes identical to ITC1013 “Sena” (AA). We then identified Genotype Groups (GGs) composed of genotypes with pairwise dissimilarities < 0.1. Eleven GGs involving AROB accessions were identified including some corresponding to known Groups of Musa (Cavendish, Maoli-Popoulu and Iholena). Six GGs involved both AROB and ITC accessions and two were composed of AROB accessions only. We also noted that the Fe’i bananas AROB010 “Bia Kaura”, AROB026 “Kauraï”, AROB029 “Koraï 1” and AROB031 “Kourai” were part of the same GG.
Diversity clustering
The NJ tree built with the edible accessions of the Musa section and their close ancestors provided a better image of the diversity collected in Bougainville (Fig. 3). While the clustering of many accessions within the AA from PNG or their link to the AAB Pacific Groups Maoli-Popoulu and Iholena was not surprising, we noted that AROB024 “Seven Kina” (AAA) was linked to the East African Mutika/Lujugira (AAA) while AROB057 “Sepik” (AAA) was located within a wide cluster of various AA and AAA mainly from SE Asia, but was not directly branching on any accession from the CS. One accession, AROB020 “Kararu”, clustered near the Laknau Group (AAB) from the Philippines. We also noticed that AROB015 “Laguai”, AROB017 “Banawa” and AROB058 “Korukapi” (all triploid AAA) clustered within the AA PNG Group originally composed of AA only. The tetraploids accession AROB027 “Buka” and AROB056 “Kalmagol” clustered within the ABB Pisang Awak Group and with the AAB Silk/AB Kunnan Groups, respectively. Four accessions branched on M. schizocarpa and clustered with two natural hybrids between M. acuminata ssp. banksii and M. schizocarpa (ITC0859 and ITC0822 “Sosi”) and a cultivated variety classified as AS, ITC0822 “Tonton Kepa”. Among these accessions, three are ornamentals (AROB001 “Flower Banana”, AROB006 “Nono 1” and AROB041 “Glenda’s Red”). The fourth one is AROB032, a triploid edible accession, named “Toitoi” by the team and originally classified as potential AAB.
Refining the classification of doubtful accessions
Combining the observations on the morphology, ploidy estimation and SSR genotyping we confirmed the classification determined in the field for quite a number of accessions and we were able to refine the classifications for the accessions for which we had doubts (Table 2). Due to its clustering with the ABB Pisang Awak Group, the genomic composition AABB is proposed for the tetraploid AROB027 “Buka”. Despite its clustering with Silk AAB, we propose to stick to Allen’s classification AABB for AROB056 “Kalmagol” which looked similar to “Kalamagol” AABB collected in Bougainville in the 1960’s (Rosales et al. 1999). AROB009 “Bukatawawe” was classified as triploid by flow cytometry and therefore is confirmed AAA. AROB032 “Toitoi” was classified as triploid and branched on M. schizocarpa therefore suggesting a genomic composition of AAS. This is consistent with the colour of young fruits observed that is similar to M. schizocarpa. If confirmed, AROB032 “Toitoi” would be, at our knowledge, the first recorded triploid composed of an S genome. AROB001 “Flower Banana”, AROB006 “Nono 1” and AROB041 “Glenda’s Red” also clustered near these hybrids, but their morphology was so different that further investigation is necessary to confirm the AS classification. AROB038 “Sinsiruai” and AROB061 “Sausage banana” were diploid and clustered within the AA from PNG and were therefore both classified as AA. As AROB057 “Sepik” was triploid and clustered within a group of AA/AAA accessions, we classified it as AAA. However, due to its peculiar morphology and to the two new alleles discovered in its genotype, we cannot exclude introgression from a genepool that is not present in the CS.
Discussion
The collecting mission to AROB was a fruitful exploration. Despite the four extensive collecting missions achieved in PNG in the 1980′s, many new cultivars were discovered.
Coupling ploidy estimation and SSR genotyping using the standardized platform for molecular characterization of Musa germplasm (Christelová et al. 2011) with the field prospections was very useful for different purposes. First, the joint analysis of the CS and of the AROB accessions helped in refining or confirming the classification of the AROB accessions by complementing the observations made in the field. The most appropriate stage for banana cultivars description and identification is when the first fruits are ripe (TAG 2010) but it was not always possible on the field to find plants at this particular stage of development. Second, for the same reason and also due to G x E interactions, the formal identification of synonyms/duplicates was not always possible. The molecular characterization of the collected accessions allowed detecting potential duplicates within the newly collected accessions but also within the joint CS-AROB datasets. It also allowed identifying nearly identical genotypes deriving from clonal diversification. Due to the accumulation of mutations and epigenetic changes, strictly identical genotypes do not always have strictly identical phenotypes, and therefore do not correspond to the same varieties, but they give a good estimation of the genetic diversity that was collected.
The only limitation we found using this set of markers is for the Callimusa accessions, which appear genetically very similar with the SSR markers used. The high rates of missing data observed overall in this section correspond to non-amplifying loci. It suggests high rates of null alleles for some of the markers used or the total absence of the site for some others such as the marker mMaCIR164 which was missing in all accessions of the section Callimusa. The transferability rate of SSR markers between species within genera in monocots was estimated 60% in average, of which only 40% are expected to be polymorphic (Barbará et al. 2007). As the SSR markers used here were developed from the Musa species M. acuminata and M. balbisiana (Crouch et al. 1998; Lagoda et al. 1998; Hippolyte et al. 2010) that belong both to the Musa section, the Callimusa species may be too genetically distant. Recently, SNPs called from the mapping of Callimusa reads on the M. acuminata reference genome were not accurate (Y. Hueber and M. Rouard pers. com.) and may also reflect high levels of differentiation. A set of markers developed specifically for targeted Callimusa species would be likely to lead to different results. Therefore, the interpretation of the results on the wild and cultivated Callimusa genotypes in this study should be made cautiously.
The clustering of some of the AROB accessions within the CS was particularly interesting with regard to the diversification history of cultivated bananas. The emergence of triploid cultivars ensued from sexual diversification through the occurrence of unbalanced meiosis leading to unreduced gametes within edible diploids (De Langhe et al. 2009; Perrier et al. 2011). For example, the AAA Cavendish clones resulted from a natural cross between two AA landraces from the Mlali Group (2n gamete donor) and “Khai Nai On” (n gamete donor) (Carreel et al. 2002; Raboin et al. 2005; Perrier et al. 2009; Hippolyte et al. 2012). In this context, the positions of the three AAA AROB015 “Laguai”, AROB017 “Banawa” and AROB058 “Korukapi” in the diversity tree are also of particular interest as they are the only polyploids observed within a wide cluster of edible AA collected in PNG and closely related to M. acuminata subsp. banksii (F.Muell.) N.W.Simmonds, the Papuan subspecies of M. acuminata. Until now, only diploids have been reported in this genetic cluster (Christelová et al. 2017; Sardos et al. 2016b) and this pattern suggests that these triploids likely resulted from natural sexual crosses within Papuan edible diploid bananas. The two tetraploid accessions collected, AROB027 “Buka” and AROB056 “Kalmagol”, are also of particular interest. Their locations in the tree and their morphologies suggest the Groups Pisang Awak (ABB) and Silk (AAB) or Kunnan (AB) as parents, respectively. None of these triploid Groups is native to the region but the Pisang Awak Group is grown worldwide. In PNG, it is notably used to produce a local alcohol named Jungle Juice. Seeds are known to be quite common in cultivars from the Pisang Awak Group in their centre of origin, Malaysia (Simmonds 1966). Given the triploid status of this Group, viable progeny would be likely resulting from the natural cross of an unreduced gamete (3n) from the mother plant with a haploid pollen grain (n gamete) from a diploid plant. The tetraploid plant obtained from such a cross could be similar in morphology to its 3x parent, such as AROB027 “Buka”, and may be wrongly classified as part of the Pisang Awak Group. This may be the origin of a debate regarding the ploidy level of the Pisang Awak Group, as some tetraploids have been reported within this Group (Pillay et al. 2006).
The occurrence of a tetraploid linked to the Silk Group (AAB) and Kunnan Group (AB), which are both from India, is more surprising. Even though AROB056 “Kalmagol” was more frequently planted than AROB027 “Buka”, we didn’t note an abundance of Silk locally, as only a single plant was observed in the town of Arawa (Sachter-Smith et al. 2017) and no diploid AB was recorded. Therefore, this tetraploid was likely introduced to the island. Allen already collected the variety under the name “Kalamagol” in the 1960′s (Rosales et al. 1999) showing it was already there 50 years ago. Given the thousands of Indian indentured labourers who were brought to the Pacific, mainly to Fiji, by the British Empire in the 19th century, AROB056 “Kalmagol” may have reached Bougainville from India via Fiji. Interestingly, “Kalamagol” sounds similar to “Kalaimagal” which is a feminine name of Indian origin.
Due to high levels of sterility, many banana varieties are clonal selections from a single clone. For example, the more than 150 known Plantain varieties are considered to be derived from the clonal diversification of a single original plant (Noyer et al. 2005). Comparing the AROB accessions and the CS, we identified six pairs and a triplet of identical genotypes. If it strongly suggests that these pairs/triplet are duplicates, it is not the case for the pair AROB023 “Morou”/AROB055 “Tambra”. AROB055 “Tambra” is indeed variegated, that is to say that its leaves and fruits exhibit white stripes, while AROB023 “Morou” is not (Sachter-Smith et al. 2017). Variegation in plants is due to the partial fixation of a mutation in the chlorophyll-synthesis pathway (Marcotrigiano 1997). It is therefore likely that AROB055 “Tambra” ensues from the clonal diversification of AROB023 “Morou” and was intentionally selected by farmers for its attractive morphology. Such a case was already documented for cassava in the Pacific archipelago of Vanuatu, located further south of the Solomon Islands, where farmers capture all the variations of an initial genotype, independently of whether these variations might be agronomically useful (Caillon and Lanouguère-Bruneau 2005; Sardos et al. 2008). Here, we therefore documented in banana the fixation of an obvious mutation followed by the intentional and likely local selection of the variant by farmers. If variegated cassava is mostly used as an ornamental plant, we observed variegated bananas sold in the market in Buka town, attesting the potential value of growing such a variety.
Conclusion
The results of the collecting mission presented in this paper confirmed PNG as an important hotspot for banana genetic diversity as new genotypes, and new alleles, were collected despite the extensive collecting missions performed in the country in the past. The results of the SSR genotyping achieved concomitantly with the mission allowed highlighting not only the diversity cultivated in the AROB but also the different diversification processes at work in the region despite the crop’s vegetative mode of propagation. The maintenance of crop evolution under farmer management is key to the successful establishment of in-situ and on -farm conservation initiatives (Bellon et al. 2017). Our results suggest the occurrence of gene flow, the accumulation of mutations and the introduction of new varieties in the AROB. Even though further studies should be undertaken in the future to fully characterize and understand these processes, we already have good insights that AROB and, by extension, PNG is an excellent candidate for the establishment of on -farm conservation programmes for Musa.
Notes
These missions were organized by IBPGR and QDPI (current Queensland DAF) in co-operation with the PNG Department of Agriculture and Livestock (current NARI) and supported by INIBAP (current Bioversity International).
Since 1994 and the signature of an agreement between Bioversity International and FAO, all the germplasm conserved in the ITC, including the PNG material, is available to all on the understanding that it remains in the public domain.
References
Argent GCG (1976) The wild bananas of Papua New Guinea. Notes R Bot Gard 35:77–114
Arnaud E, Horry JP (1997) Musalogue: a catalogue of Musa germplasm. Papua New Guinea collecting missions, 1988–1989. INIBAP, Montpellier
Ball T, Vrydaghs L, Van Den Hauwe I, Manwaring J, De Langhe E (2006) Differentiating banana phytoliths: wild and edible Musa acuminata and Musa balbisiana. J Archaeol Sci 33:1228–1236
Barbará T, Palma-Silva C, Paggi GM, Bered F, Fay MF, Lexer C (2007) Cross-species transfer of nuclear microsatellite markers: potential and limitations. Mol Ecol 16:3759–3767. https://doi.org/10.1111/j.1365-294X.2007.03439.x
Bardou P, Mariette J, Escudié F, Djemiel C, Klopp C (2014) jvenn: an interactive Venn diagram viewer. BMC Bioinf 15:293. https://doi.org/10.1186/1471-2105-15-293
Bartoš J, Alkhimova O, Dolezelova M, De Langhe E, Dolezel J (2005) Nuclear genome size and genomic distribution of ribosomal DNA in Musa and Ensete (Musaceae): taxonomic implications. Cytogenet Genome Res 109:50–57. https://doi.org/10.1159/000082381
Bellon MR, Dulloo E, Sardos J, Thormann I, Burdon JJ (2017) In situ conservation—harnessing natural and human-derived evolutionary forces to ensure future crop adaptation. Evol Appl 00:1–13. https://doi.org/10.1111/eva.12521
Bourke RM, Woruba M, Allen BJ, Allen M, Grau R, Hobsbawn P (2002) Bougainville province: text summaries, maps, code lists and village identification. Agricultural systems of Papua New Guinea working paper no. 20. Land Management Group, Department of Human Geography, Research School of Pacific and Asian Studies, The Australian National University, Canberra. Revised edition
Caillon S, Lanouguère-Bruneau V (2005) Ethnoécologie en Océanie: gestion de l’agrobiodiversité dans un village de Vanua Lava (Vanuatu): stratégies de sélection et enjeux sociaux. J Soc Ocean 120:129–148
Carreel F, Gonzales De Léon D, Lagoda P, Lanaud C, Jenny C, Horry J, Tézenas du Montcel H (2002) Ascertaining maternal and paternal lineage within Musa by chloroplast and mitochondrial DNA RFLP analyses. Genome 45:679–692. https://doi.org/10.1139/g02-033
Christelová P, Valárik M, Hřibová E, Van den Houwe I, Channelière S, Roux N, Doležel J (2011) A platform for efficient genotyping in Musa using microsatellite markers. AoB Plants 2011:plr024
Christelová P, De Langhe E, Hřibová E, Čížková J, Sardos J, Hušáková M, Van Den Houwe I, Sutanto A, Kepler AK, Swennen R, Roux N, Doležel J (2017) Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers Conserv 26:801–824. https://doi.org/10.1007/s10531-016-1273-9
Čížková J, Hřibová E, Christelová P, Van Den Houwe I, Häkkinen M, Roux N, Swennen R, Doležel J (2015) Molecular and cytogenetic characterization of wild Musa species. PLoS ONE 10(8):e0134096. https://doi.org/10.1371/journal.pone.0134096
Crouch HK, Crouch JH, Jarret R, Cregan PB, Ortiz R (1998) Segregation at microsatellite loci in haploid and diploid gametes of Musa. Crop Sci 38:211–217
De Langhe E, Vrydaghs L, De Maret P, Perrier X, Denham TP (2009) Why bananas matter: an introduction to the history of banana domestication. Ethnobot Res Appl 7:165–177. http://journals.sfu.ca/era/index.php/era/article/view/356
Doležel J, Lysák MA, Van den houwe I, Doleželová M, Roux N (1997) Use of flow cytometry for rapid ploidy determination in Musa species. InfoMusa 6:6–9
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2(9):2233–2244. https://doi.org/10.1038/nprot.2007.310
Douhovnikoff V, Dodd RS (2003) Intra-clonal variation and a similarity threshold for identification of clones: application to Salix exigua using AFLP molecular markers. Theor Appl Genet 106(7):1307–1315. https://doi.org/10.1007/s00122-003-1200-9
FAO (2014) http://www.fao.org/3/a-i4036e.pdf
Galbraith DW, Doležel J, Lambert G, Macas J (1998) Nuclear DNA content and ploidy analyses in higher plants. In: Robinson JP (ed) Current protocols in cytometry. Wiley, New York, pp 7.6.1–7.6.22
Geering ADW, Olszewski NE, Harper G, Lockhart BEL, Hull R, Thomas JE (2005) Banana contains a diverse array of endogenous badnaviruses. J Gen Virol 86:511–520
Häkkinen M (2013) Reappraisal of sectional taxonomy in Musa (Musaceae). Taxon 62(4):809–813. https://doi.org/10.12705/624.3
Heslop-Harrison JS, Schwarzacher T (2007) Domestication, genomics and the future for banana. Ann Bot 100(5):1073–1084. https://doi.org/10.1093/aob/mcm191
Hippolyte I, Bakry F, Séguin M, Gardes L, Rivallan R, Risterucci AM, Jenny C, Perrier X, Carreel F, Argout X, Piffanelli P, Khan I, Miller RN, Pappas GJ, Mbeguie-A-Mbeguie D, Matsumoto T, de Bernardinis V, Huttner E, Kilian A, Baurens FC, D’Hont A, Côte F, Courtois B, Glaszmann JC (2010) A saturated SSR/DArT linkage map of Musa acuminata addressing genome rearrangements among bananas. BMC Plant Biol 10:65. https://doi.org/10.1186/1471-2229-10-65
Hippolyte I, Jenny C, Gardes L, Bakry F, Rivallan R, Pomies V, Cubry P, Tomekpe K, Risterucci AM, Roux N, Rouard M, Arnaud E, Kolesnikova-Allen M, Perrier X (2012) Foundation characteristics of edible Musa triploids revealed from allelic distribution of SSR markers. Ann Bot-Lond 109:937–951. https://doi.org/10.1093/aob/mcs010
Hřibová E, Čížková J, Christelová P, Taudien S, de Langhe E, Doležel J (2011) The ITS1-5.8S-ITS2 sequence region in the Musaceae: structure, diversity and use in molecular phylogeny. PLoS ONE 6:e17863
Janssens SB, Vandelook F, De Langhe E, Verstraete B, Smets E, Vandenhouwe I, Swennen R (2016) Evolutionary dynamics and biogeography of Musaceae reveal a correlation between the diversification of the banana family and the geological and climatic history of Southeast Asia. New Phytol 210:1453–1465. https://doi.org/10.1111/nph.13856
Lagoda PJL, Noyer JL, Dambier D, Baurens F, Grapin Lanaud C (1998) Sequence tagged microsatellite site (STMS) markers in the Musaceae. Mol Ecol 7:659–663
Lebot V (1999) Biomolecular evidence for plant domestication in Sahul. Gen Res Crop Evol 46:619–628
Marcotrigiano M (1997) Chimeras and variegation: pattern of deceit. HortScience 32:773–784
Noyer JL, Causse S, Tomekpe K, Bouet A, Baurens FC (2005) A new image of plantain diversity assessed by SSR, AFLP and MSAP markers. Genetica 124:61–69. https://doi.org/10.1007/s10709-004-7319-z
Ortiz R (2013) Conventional banana and plantain breeding. In: Van den Bergh I, Amorim EP, Johnson V (eds) Proceedings of international ISHS-promusa symposium on bananas and plantains: towards sustainable global production and improved use, Salvador (Bahia), Brazil, 2011/10/10-14. Acta Horticulturae 986. ISHS, Leuven, p 177–194
Otto F (1990) DAPI staining of fixed cells for high-resolution flow cytometry of nuclear DNA. In: Crissman HA, Darzynkiewicz Z (eds) Methods in cell biology, vol 33. Academic Press, New York, pp 105–110
Perrier X, Jacquemoud-Collet JP (2006) DARwin software version 6.0. CIRAD. http://www.darwin.cirad.fr/darwin
Perrier X, Flori A, Bonnot F (2003) Data analysis methods. In: Hamon P, Seguin M, Perrier X, Glaszmann JC (eds) Genetic diversity of cultivated tropical plants. Sciences Publisher, Montpellier, pp 43–76
Perrier X, Bakry F, Carreel F, Jenny C, Horry JP, Lebot V, Hippolyte I (2009) Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot Res Appl 7:199–216. http://journals.sfu.ca/era/index.php/era/article/view/362
Perrier X, De Langhe E, Donohue M, Lentfer C, Vrydaghs L, Bakry F, Carreel F, Hippolyte I, Horry JP, Jenny C, Lebot V, Risterucci AM, Tomekpe K, Doutrelepont H, Ball T, Manwaring J, De Maret P, Denham T (2011) Multidisciplinary perspectives on banana (Musa spp.) domestication. PNAS 108:11311–11318. https://doi.org/10.1073/pnas.1102001108
Pillay M, Oqundiwan E, Tenkouano A, Doležel J (2006) Ploidy and genome composition of Musa germplasm at the International Institute of Tropical Agriculture. Afr J Biotechnol 5:1224–1232
Ploetz RC, Haynes JL, Vázquez A (1999) Responses of new banana accessions in South Florida to Panama disease. Crop Prot 18:445–449
Raboin LM, Carreel F, Noyer JL, Baurens FC, Horry JP, Bakry F, Du Montcel HT, Ganry J, Lanaud C, Lagoda PJL (2005) Diploid ancestors of triploid export banana cultivars: molecular identification of 2n restitution gamete donors and n gamete donors. Mol Breeding 16:333–341
Rosales FE, Arnaud E, Coto J (1999) A tribute to the work of Paul Allen: a catalogue of wild and cultivated bananas. INIBAP, Montpellier
Sachter-Smith G, Paofa J, Rauka G, Sardos J, Janssens S (2017) Bananas of The Autonomous Region of Bougainville: a catalog of banana diversity seen on the islands of Bougainville and Buka, Papua New Guinea. Bioversity International, Montpellier
Sardos J, McKey D, Duval M-F, Malapa R, Noyer J-L, Lebot V (2008) Evolution of cassava (Manihot esculenta Crantz) after recent introduction into a South Pacific island system: the contribution of sex to the diversification of a clonally propagated crop. Genome 51:912–921
Sardos J, Rouard M, Hueber Y, Cenci A, Hyma KE, van Den Houwe I, Hřibová E, Courtois B, Roux N (2016a) A genome-wide association study on the seedless phenotype in banana (Musa spp.) reveals the potential of a selected panel to detect candidate genes in a vegetatively propagated crop. PLoS ONE 11:e0154448
Sardos J, Perrier X, Doležel J, Hřibová E, Christelová P, Van den Houwe I, Kilian A, Roux N (2016b) DArT whole genome profiling provides insights on the evolution and taxonomy of edible Banana (Musa spp.). Ann Bot-Lond 118:1269–1278. https://doi.org/10.1093/aob/mcw170
Simmonds NW (1966) Bananas, 2nd edn. Tropical Agricultural Series. Longman, New York
Taxonomy Advisory Group (TAG) (2010) Minimum descriptor list for Musa. Revised 2016. Bioversity International, Montpellier
Tenkouano A, Pillay M, Ortiz R (2011) Breeding techniques. In: Pillay M, Tenkouano A (eds) Banana breeding progress and challenges. CRC Press, Boca Raton, pp 181–200
Teo CH, Pui HP, Othman RY, Harikrishna JA (2011) Comparative analysis of Argonaute gene sequences in bananas (Musa sp.) shows conserved species-specific Ago-7 PIWI domains. Gen Res Crop Evol 58:713–725
Till BJ, Jankowicz-Cieslak J, Sági L, Huynh OA, Utsushi H, Swennen R, Terauchi R, Mba C (2010) Discovery of nucleotide polymorphisms in the Musa gene pool by ecotilling. Theor Appl Genet 121:1381–1389
Tomekpe K, Jenny C, Escalant J (2004) A review of conventional improvement strategies for Musa. Infomusa 13:2–6
Valdez-Ojeda R, James-Kay A, Ku-Cauich JR, Escobedo-Gracia Medrano RM (2014) Genetic relationships among a collection of Musa germplasm by fluorescent-labeled SRAP tree. Genet Genomes 10:465–476
Volkaert H (2011) Molecular analysis reveals multiple domestications of edible bananas. In: Van den Bergh I, Smith M, Swennen R, Hermanto C (eds.) Proceedings of international ISHS-ProMusa symposium on global perspectives on Asian challenges. Acta Horticulturae 897, ISHS, Leuven, pp 143–152
Acknowledgements
In the Autonomous Region of Bougainville, we wish to thank Mr. Zohn Bosco Miriona from Bougainville Express Tout (BET) for being our guide during the whole mission and the Department of Primary Industry officers from Buka, Kieta and Buin for their support. Finally, we acknowledge local farmers who kindly accepted to provide samples of their banana varieties and answered our questions on their origin and use. Many thanks to Rachel Chase of Bioversity International for editing this manuscript. This work was funded by the Genebank CGIAR Research Programme and the CGIAR Research Programme on Roots, Tubers and Bananas (RTB) with a contribution of the Belgium government through the PhenSeeData project. This work is also supported by the CGIAR Fund Donors. P.C., J.Č and J.D. were supported by the Czech Ministry of Education, Youth and Sports (award LG15017 and LO1204 from the National Program of Sustainability I).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10722_2018_690_MOESM1_ESM.pptx
Figure S1: Distribution of the dissimilarities calculated following the Dice index and obtained for the merged pruned CS-AROB dataset. Graph shows the first 50 classes of distance (step = 0.02) (PPTX 127 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sardos, J., Christelová, P., Čížková, J. et al. Collection of new diversity of wild and cultivated bananas (Musa spp.) in the Autonomous Region of Bougainville, Papua New Guinea. Genet Resour Crop Evol 65, 2267–2286 (2018). https://doi.org/10.1007/s10722-018-0690-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-018-0690-x