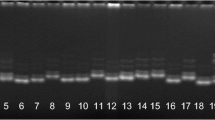
Abstract
Morphological traits and two kinds of molecular markers were employed to study the genetic relationships among improved rice (Oryza sativa ) varieties of Indonesia since 1943. Dendrograms based on morphological traits and both molecular markers (simple sequence repeats, SSR and single nucleotide polymorphism, SNP) agreed in separating the varieties into two primary groups. Based on the morphological traits, a larger group (>60 %) contains varieties with smaller sizes compared with those in the smaller group (<40 %). SSR and SNP markers revealed that most of the varieties belonged to indica (88; 89 %) and japonica (9; 8 %) subspecies, and 3 % of varieties were not involved in two subspecies. The molecular markers revealed that the genetic diversity (H) stagnated between stage II (1967–1985) and stage III (1986–2003). However, during stage I (1943–1966), H was higher than in the other stages as revealed by SNP markers, while H in stage I was lower than in the other stages as revealed by SSR markers. In this study, the two molecular data sets were positively correlated and positive correlations between the phenotypic and molecular data depended on the kind of molecular marker: SNP had higher Mantel r values than SSRs. Besides, SSR markers seem to be appropriate for pedigree studies, while SNP markers could be used to reveal genomic relationships. These findings were attributable to the different properties of these two different markers. These results suggested that the diversity and differentiation of both the phenotypic and molecular marker variations were probably resulted from the crossing and selection in rice breeding in Indonesia. We suggest that Indonesia needs another strategy to improve new varieties to avoid a reduction in genetic diversity and similarity.






Similar content being viewed by others
References
Caldo RA, Sebastian LS, Hernandez JE (1996) Multivarieate analysis of phenotypic diversity of improved rice varieties. Philipp J Crop Sci 21:76–85
Choudhary G, Ranjitkumar N, Surapaneni M, Deborah DA, Vipparla A, Anuradha G, Siddiq EA, Vemireddy LR (2013) Molecular genetic diversity of major Indian rice cultivars over decadal periods molecular genetic diversity of major Indian rice cultivars over decadal periods. PLoS One 8:e66197
Chunhai S, Zongtan S (1995) Tall and dwarf near isogenic lines of indica rice characterized. Int Rice Res Notes 20:4–6
CIRAD (2009) Dissimilarity analysis and representation for windows. http://darwin.cirad.fr/darwin/Home.php
Crowell S, Falcão AX, Shah A, Wilson Z, Greenberg AJ, McCouch SR (2014) High-resolution inflorescence phenotyping using a novel image-analysis pipeline, PANorama. Plant Physiol 165:479–495
Diederichsen A (2009) Duplication assessments in Nordic Avena sativa accessions at the Canadian national genebank. Genet Resour Crop Evol 56:587–597
Emanuelli F, Lorenzi S, Grzeskowiak L, Catalano V, Stefanini M, Troggio M, Myles S, Martinez-Zapater JM, Zyprian E, Moreira FM, Grando MS (2013) Genetic diversity and population structure assessed by SSR and SNPs in a large germplasm collection of grape. BMC Plant Biol 13:1–17
Foll M, Gaggiotti OE (2008) A genome scans method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Fuentes JL, Cornide MT, Alvarez A, Suarez E, Borges E (2005) Genetic diversity analysis of rice varieties (Oryza sativa L.) based on morphological, pedigree and DNA polymorphism data. Plant Genet Resour 3:353–359
Ganal MW, Altmann T, Röder MS (2009) SNP identification in crop plants. Curr Opin Plant Biol 12:211–217
Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch SR (2005) Genetic structure and diversity in Oryza sativa L. Genetics 169:1631–1638
Glaszmann JC (1987) Isozymes and classification of Asian rice varieties. Theor Appl Genet 74:21–30
Hamblin MT, Warburton ML, Buckler ES (2007) Empirical comparison of simple sequence repeats and single nucleotide polymorphisms in assessment of maize diversity and relatedness. PLoS One 2:e1367
Hargrove TR, Cabanilla VL, Coffman WR (1988) Twenty years of rice breeding. Bioscience 38:675–681
Hossain M (2007) Rice facts: a balancing act. Rice Today 6:37
IBPGR-IRRI (1980) Descriptor for rice Oryza sativa L. IRRI, Manila, pp 6–21
Jenkins S, Gibson N (2002) High-throughput SNP genotyping. Comp Funct Genom 3:57–66
JMP® (2000) Version 5. SAS Institute Inc, Cary
Khush GS (1997) Origin, dispersal, cultivation and variation of rice. Plant Mol Biol 35:25–34
Kunusoth K, Vadivel K, Sundaram RM, Sultana R, Rajendrakumar P, Maganti S, Subbarao LV, Chin-Wo SR (2015) Assessment of genetic diversity of elite Indian rice varieties using agro-morphological traits and SSRs. Am J Exp Agric 6:384–401
Li ZK, Yu SB, Lafitte HR, Huang N, Courtois B, Hittalmani S, Vijayakumar CHM, Liu GF, Wang GC, Shashidhar HE, Zhuang JY, Zheng KL, Singh VP, Sidhu JS, Srivantaneeyakul S, Khush GS (2003) QTL environment interactions in rice. I. Heading date and plant height. Theor Appl Genet 108:141–153
Lu H, Redus MA, Coburn JR, Rutger JN, McCouch SR, Tai TH (2005) Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR Marker analysis. Crop Sci 45:66–76
Mantegazza R, Biloni M, Grassi F, Basso B, Lu B-R, Cai X-X, Sala F, Spada A (2008) Temporal trends of variation in Italian rice germplasm over the past two centuries revealed by AFLP and SSRs. Crop Sci 48:1832–1840
McCouch SR, Kochert G, Yu ZH, Wang ZY, Khush GS, Coffman WR, Tanksley SD (1988) Molecular mapping of rice chromosomes. Theor Appl Genet 76:815–829
McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, Stokowskih R, Ballingerh DG, Frazerh KA, Coxh DR, Padhukasahasrame B, Bustamantee CD, Weigelf D, Mackilla DJ, Bruskiewicha RM, Ratschc G, Buellb CR, Leunga H, Leach JE (2009) Genome wide SNP variation reveals relationships among landraces and modern varieties of rice. PNAS 106:12273–12278
Muhamad K, Ebana K, Fukuoka S, Okuno K (2015) Diversity and differentiation of Oryza sativa and O. rufipogon in Indonesia. Genet Resour Crop Evol 62:1–14
Murai M, Takamure I, Sato S, Tokutome T, Sato Y (2002) Effects of the dwarfing gene originating from ‘Dee-geo-woo-gen’ on yield and its related traits in rice breeding. Science 52:95–100
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Ogunbayo SA, Ojo DK, Guei RG, Oyelakin OO, Sanni KA (2005) Phylogenetic diversity and relationships among 40 rice accessions using morphological and RAPDs techniques. Afr J Biotechnol 4:1234–1244
Peakall R, Peter ES (2006) GENALEX 6: genetic analysis in excel population genetic software for teaching and research. Mol Ecol 6:288–295
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) Green revolution genes code mutant gibberellin response modulators. Nature 400:256–261
Perrier X, Jacquemod-Collet JP (2006) DARwin software. http://darwin.cirad.fr/
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system, version 2.2. Exeter Software, Setauket
Sasaki A, Ashikari M, Tanaka MU, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M (2002) Green revolution: a mutant gibberellins-synthesis gene in rice. Nature 416:701–702
Shannon CA (1948) Mathematical theory of communication. Bell Syst Technol J 27:379–423
Spada A, Mantegazza R, Biloni M, Caporali E, Sala F (2004) Italian rice varieties: historical data, molecular markers and pedigrees to reveal their genetic relationships. Plant Breed 123:105–111
Staub JE (1994) Crossover: concepts in applications in genetics, evolution and breeding—an interactive computer-based laboratory manual. University Of Wisconsin Press, Wisconsin
Susanto U, Daradjat AA, Suprihatno B (2003) Advance in lowland rice breeding in Indonesia. J Litbang Pertan 22:125–131 (abstract in English)
Tanaka Y, Shiraiwa T (2009) Stem growth habit affects leaf morphology and gas exchange traits in soybean. Ann Bot 104:1293–1299
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Thomson MJ, Septiningsih EM, Suwardjo F, Santoso TJ, Silitonga TS, McCouch SR (2007) Genetic diversity analysis of traditional and improved Indonesian rice (Oryza sativa L.) germplasm using microsatellite markers. Theor Appl Genet 114:559–568
Tsai KH (1998) Effects of genes controlling plant height in near isogenic lines of indica and japonica cultivars. RGN 15:103–104
UNCTAD (2011) Historical evolution of the main paddy rice consuming countries (human food) in tonnes between 1961 and 2002. http://www.unctad.info/en/Infocomm/Agricultural_Products/Riz/Market/. Accessed 2 Mar 2015
Vergara BS, Chang TT (1985) The flowering response of the rice plant to photoperiod: a review of the literature, 4th edn. IRRI, Manila
Vieira FG, Fumagalli M, Albrechtsen A, Nielsen R (2013) Estimating inbreeding coefficients from NGS data: impact on genotype calling and allele frequency estimation. Genome Res 23:1852–1861
Wei X, Yuan X, Yu H, Wang Y, Xu Q, Tang S (2009) Temporal changes in SSR allelic diversity of major rice cultivars in China. J Genet Genom 36:363–370
Yonemaru J-I, Ebana K, Yano M (2014) HapRice, an SNP haplotype database and a web tool for rice. Plant Cell Physiol 55:e9 (1–12)
Yuan X-P, Wei X-H, Hua L, Yu H-Y, Wang Y-P, Xu Q, Tang S-X (2007) A somparative study of SSRDiversity in Chinese major rice varieties planted in 1950s and in the recent ten years (1995–2004). Rice Sci 14:78–84
Yue B, Xue W, Luo L, Xing YJ (2008) Identification of quantitative trait loci for four morphologic traits under water stress in rice (Oryza sativa L.). J Genet Genom 35:569–575
Zhang P, Li J, Li X, Liu X, Zhao X, Lu Y (2011) Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSRs. PLoS One 6:e27565
Acknowledgments
We are grateful for the Dikti Scholarship and would like to thank the Government of the Indonesian Republic, who funded the first author for a Ph.D. program at the University of Tsukuba, and the ICRR for providing us seed material for this study.
Authors’ contributions
The first author contributes to substantive conception, laboratory and field work, design, analysis, interpretation of data and drafts the manuscript; the second, the third and the fourth authors contributed to revise it critically for important intellectual content; and the fourth author also as a final approval person of the version to be published.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Muhamad, K., Ebana, K., Fukuoka, S. et al. Genetic relationships among improved varieties of rice (Oryza sativa L.) in Indonesia over the last 60 years as revealed by morphological traits and DNA markers. Genet Resour Crop Evol 64, 701–715 (2017). https://doi.org/10.1007/s10722-016-0392-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-016-0392-1