Abstract
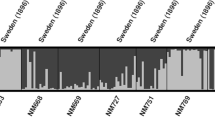
Landrace accessions have long been recognized as an important source of genetic diversity for crop species, and landraces are stored in genebanks across the world as genetic resources for future crop development. Landraces are also an important part of the human cultural heritage and as such they have been used for genetic studies to make inferences about historical agriculture. However, surprisingly little is known about the within-accession diversity of landrace crops of different species. In order to evaluate the diversity of Swedish landraces we used microsatellite markers to genotype accessions of four species (barley, pea, oats and rye), both extant genebank material and 114-year-old seed samples of similar geographic origin and type. We found consistently high levels of within-population genetic diversity in the historical material, but varying and often lower diversity levels in the genebank accessions. We also make tentative conclusions about how representative the genebank material is to what was originally cultivated in its reported area of origin and suggest that the true identity of the genebank accessions is unclear and that historical seed collections should be a more appropriate material for the study of historical agriculture.




Similar content being viewed by others
References
Abdel-Ghani AH, Parzies HK, Omary A, Geiger HH (2004) Estimating the outcrossing rate of barley landraces and wild barley populations collected from ecologically different regions of Jordan. Theor Appl Genet 109:588–595
Atterberg A (1891) Neues System der Hafervarietäten nebst Beschreibung der nordischen Haferformen. Landw Versuchs-Stat 39:171–204
Camacho Villa TC, Maxted N, Scholten MA, Ford-Lloyd BV (2005) Defining and identifying crop landraces. Plant Gen Res 3:373–384
Chebotar S, Röder MS, Korzun V, Börner A (2002) Genetic integrity of ex situ genebank collections. Cell Mol Biol Lett 7:437–444
Chebotar S, Röder MS, Korzun V, Saal B, Weber WE, Börner A (2003) Molecular studies on genetic integrity of open-pollinating species rye (Secale cereale L.) after long-term genebank maintenance. Theor Appl Genet 107:1469–1476
Djè Y, Forcioli D, Ater M, Lefèbvre C, Vekemans X (1999) Assessing population genetic structure of sorghum landraces from North-western Morocco using allozyme and microsatellite markers. Theor Appl Genet 99:157–163
Dreisigacker S, Zhang P, Warburton ML, Skovmand B, Hoisington D, Melchinger AE (2005) Genetic diversity among and within CIMMYT wheat landrace accessions investigated with SSRs and implications for plant genetic resources management. Crop Sci 45:653–661
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fikiru E, Tesfaye K, Bekele E (2007) Genetic diversity and population structure of Ethiopian lentil (Lens culinaris Medikus) landraces as revealed by ISSR marker. Afr J Biotechnol 6:1460–1468
Harlan JR (1975) Our vanishing genetic resources. Science 188:618–621
Isaac AD, Muldoon M, Brown KA, Brown TA (2010) Genetic analysis of wheat landraces enables the location of the first agricultural sites in Italy to be identified. J Arch Sci 37:950–956
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jana S, Pietrzak LN (1988) Comparative assessment of genetic diversity in wild and primitive cultivated barley in a center of diversity. Genetics 119:981–990
Jensen NF (1961) Genetics and inheritance in oats: inheritance of morphological and other characters. In: Coffman FA (ed) Oats and oats improvement. American Society of Agronomy, Madison, WI, pp 125–136
Jones H, Lister DL, Bower MA, Leigh FJ, Smith LMJ, Jones MK (2008a) Approaches and constraints of using existing landraces and extant plant material to understand agricultural spread in prehistory. Plant Gen Res 6:98–112
Jones H, Leigh FJ, Mackay I, Bower MA, Smith LM, Charles MP, Jones G, Jones MK, Brown TA, Powell W (2008b) Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol 25:2211–2219
Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, Lundqvist U, Fujimura T, Matsuoka M, Matsumoto T, Yano M (2007) Six-rowed barley originated from a mutation in a homeodomain-leucine zipper I-class homeobox gene. Proc Natl Acad Sci 104:1424–1429
Leino MW, Hagenblad J (2010) Nineteenth century seeds reveal the population genetics of landrace barley (Hordeum vulgare). Mol Biol Evol 27:964–973
Leino MW, Hagenblad J, Edqvist J, Karlsson Strese E-M (2009) DNA preservation and utility of a historic seed collection. Seed Sci Res 19:125–135
Li CD, Rossnagel BG, Scoles GJ (2000) The development of oat microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor Appl Genet 101:1259–1268
Lia VV, Confalonieri VA, Ratto N, Cámara Hernández JA, Miante Alzogaray AM, Poggio L, Brown TA (2007) Microsatellite typing of ancient maize: insights into the history of agriculture in Southern America. Proc R Soc B 274:545–554
Lister DL, Thaw S, Bower MA, Jones H, Charles MP, Jones G, Smith LMJ, Howe CJ, Brown TA, Jones MK (2009) Latitudinal variation in a photoperiod response gene in European barley: insight into the dynamics of agricultural spread from ‘historic’ specimens. J Arch Sci 36:1092–1098
Loridon K, McPhee K, Morin J, Dubreuil P, Pilet-Nayel ML, Aubert G, Rameau C, Baranger A, Coyne C, Lejeune-Hènaut Burstin J (2005) Microsatellite marker polymorphism and mapping in pea (Pisum sativum L.). Theor Appl Genet 111:1022–1031
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci 70:3321–3323
Nordic Gene Bank (1999) Nordic Gene Bank 1979–1999. Nordic Gene Bank publications 35
Olsson G (1997) Gamla lantsorter—utnyttjande och bevarande. In: Olsson G (ed) Den svenska växtförädlingens historia. KSLA, Stockholm, pp 121–130
Ortiz R, Nurminiemi M, Madsen S, Rognli OA, Bjørnstad Å (2002) Cultivar diversity in Nordic spring barley breeding (1930–1991). Euphytica 123:111–119
Osvald H (1959) Åkerns nyttoväxter. Sv. litteratur, Stockholm
Pal N, Sandhu JS, Domier LL, Kolb FL (2002) Development and characterization of microsatellite and RFLP-derived PCR markers in oat. Crop Sci 42:912–918
Papa R, Attene G, Barcaccia G, Ohgata A, Konishi T (1998) Genetic diversity in landrace populations of Hordeum vulgare L. from Sardinia, Italy, as revealed by RAPDs, isozymes and morphophenological traits. Plant Breed 117:523–530
Parzies HK, Spoor W, Ennos RA (2000) Genetic diversity of barley landrace accessions (Hordeum vulgare ssp. vulgare) conserved for different lengths of time in ex situ gene banks. Heredity 84:476–486
Perez de la Vega MP, Allard RW (1984) Mating system and genetic polymorphism in populations of Secale cereale and Secale vavilovii. Can J Genet Cytol 26:308–317
Persson K, von Bothmer R (2002) Genetic diversity amongst landraces of rye (Secale cereale L.) from northern Europe. Hereditas 136:29–38
Persson K, von Bothmer R, Gullord M, Gunnarsson E (2006) Phenotypic variation and relationships in landraces and improved varieties of rye (Secale cereale L.) from northern Europe. Gen Res Crop Evol 53:857–866
Polowick PL, Vandenberg A, Mahon JD (2002) Field assessment of outcrossing from transgenic pea (Pisum sativum L.) plants. Transgenic Res 11:515–519
Pressoir G, Berthaud J (2004) Patterns of population structure in maize landraces from the Central Valleys of Oaxaca in Mexico. Heredity 92:88–94
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2007) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org. ISBN 3-900051-07-0
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Sanchez JJ, Goodman MM, Stuber CW (2000) Isozymatic and morphological diversity in the races of maize in Mexico. Econ Bot 54:43–59
Sonnante G, Stockton T, Nodari RO, Becerra Velásquez VL, Gepts P (1994) Evolution of genetic diversity during the domestication of common-bean (Phaseolus vulgaris L.). Theor Appl Genet 89:629–635
Struss D, Plieske J (1998) The use of microsatellite markers for detection of genetic diversity in barley populations. Theor Appl Genet 97:308–315
Sun CQ, Wang XK, Li ZC, Yoshimura A, Iwata N (2001) Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor Appl Genet 102:157–162
Varshney RK, Marcel TC, Ramsay L, Russel J, Röder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A (2007) A high density barley microsatellite consensus map with 775 SSR loci. Theor Appl Genet 114:1091–1103
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323–354
Xu DH, Gai JY (2003) Genetic diversity of wild and cultivated soybeans growing in China revealed by RAPD analysis. Plant Breed 122:503–506
Zeven AC (1998) Landraces: a review of definitions and classifications. Euphytica 104:127–139
Acknowledgments
The authors would like to thank the Nordic Genetic Resource Center (NordGen) for providing seed material. This work was supported by the Lagersberg foundation, the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS), Carl Tryggers Foundation, the Nilsson-Ehle foundation and the Helge Ax:son Johnsson foundation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hagenblad, J., Zie, J. & Leino, M.W. Exploring the population genetics of genebank and historical landrace varieties. Genet Resour Crop Evol 59, 1185–1199 (2012). https://doi.org/10.1007/s10722-011-9754-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-011-9754-x