Abstract
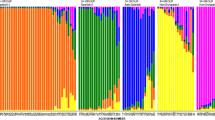
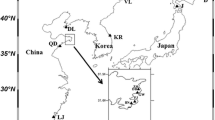
Genetic variability within and among eight landrace populations of tea (Camellia sinensis (L.) O. Kuntze) located in southern Kyoto, Japan, was surveyed with six microsatellite markers. The average number of alleles per locus was 3.83 to 4.67 for landrace populations, whereas the corresponding value among modern cultivars and breeding lines was 6.63. Expected heterozygosity values averaged over loci within landrace populations ranged from 0.498 to 0.723. A similar level of variation, 0.682, was observed for cultivars and breeding lines. High fixation index values (0.177–0.417) for each population are consistent with biparental inbreeding within the population. Genetic differentiation among local populations was extremely low with F ST = 0.062, although AMOVA revealed significant differentiation among landrace populations. We propose that these populations share a common ancestral gene pool and that some degree of artificial selection within each population has been performed by local farmers. Neighbor-joining analysis revealed that genetic relationships among populations reflect geographical location of populations. This might result from more frequent genetic exchange by nearby farmers.


Similar content being viewed by others
References
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haploytpes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Felsenstein J (2004) PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
Freeman S, West J, James C, Lea V, Mayes S (2004) Isolation and characterization of highly polymorphic microsatellites in tea (Camellia sinensis). Mol Ecol Notes 4:324–326
Fu Y-B, Diederichsen A, Richards KW, Peterson G (2002) Genetic diversity within a range of cultivars and landraces of flax (Linum usitatissimum L.) as revealed by RAPDs. Genet Res Crop Evol 49:167–174
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html
Kaundun SS, Matsumoto S (2002) Heterologous nuclear and chloroplast microsatellite amplification and variation in tea, Camellia sinensis. Genome 45:1041–1048
Kaundun SS, Matsumoto S (2003) Development of CAPS markers based on three key genes of the phenylpropanoid pathway in Tea, Camellia sinensis (L.) O. Kuntze, and differentiation between assamica and sinensis varieties. Theor Appl Genet 106:375–383
Kaundun SS, Park Y-G (2002) Genetic structure of six Korean tea populations as revealed by RAPD-PCR markers. Crop Sci 42:594–601
Kaundun SS, Zhyvoloup A, Park Y-G (2000) Evaluation of the genetic diversity among elite tea (Camellia sinensis var. sinensis) accessions using RAPD markers. Euphytica 115:7–16
Matsumoto S, Takeuchi A, Hayatsu M, Kondo S (1994) Molecular cloning of phenylalanine ammonia-lyase cDNA and classification of varieties and cultivars of tea plants (Camellia sinensis) using the tea PAL cDNA probe. Theor Appl Genet 89:671–675
Matsumoto S, Kiriiwa Y, Yamaguchi S (2004) The Korean tea plant (Camellia sinensis): RFLP analysis of genetic diversity and relationship to Japanese tea. Breed Sci 54:231–237
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Ogi Y, Ueda K, Tsuchihashi M (1995) Characterization of tea landraces in Uji, Kyoto. Tea Res J 82 (suppl.): 40–41 (in Japanese)
Park Y-G, Kaundun SS, Zhyvoloup A (2002) Use of the bulked genomic DNA-based RAPD methodology to assess the genetic diversity among abandoned Korean tea plantations. Genet Resour Crop Evol 49:159–165
Paul S, Wachira FN, Powell W, Waugh R (1997) Diversity and genetic differentiation among populations of Indian and Kenyan tea (Camellia sinensis (L.) O. Kuntze) revealed by AFLP markers. Theor Appl Genet 94:255–263
Pradeepkumar T, Karihaloo JL, Archak S, Baldev A (2003) Analysis of genetic diversity in Piper nigrum L. using RAPD markers. Genet Resour Crop Evol 50:469–475
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Shoaib A, Arabi MIE (2006) Genetic diversity among Syrian cultivated and landraces wheat revealed by AFLP markers. Genet Resour Crop Evol 53:901–906
Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) version 4. Sinauer Associates, Sunderland, MA
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ohsako, T., Ohgushi, T., Motosugi, H. et al. Microsatellite variability within and among local landrace populations of tea, Camellia sinensis (L.) O. Kuntze, in Kyoto, Japan. Genet Resour Crop Evol 55, 1047–1053 (2008). https://doi.org/10.1007/s10722-008-9311-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-008-9311-4