Abstract
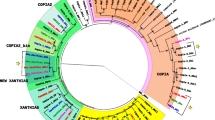
The presence of the micropia retroelement from the Ty1-copia family of LTR retroelements was investigated in three species of the Drosophila cardini group. Southern blot analysis suggested the existence of at least four micropia copies in the genomes of D. cardinoides, D. neocardini and D. polymorpha populations. The high sequence similarity between dhMiF2 and Dm11 clones (micropia retroelements isolated from D. hydei and D. melanogaster, respectively) with micropia sequences amplified from D. cardini group genome supports the hypothesis that this retroelement plays an active role in horizontal transfer events between D. hydei and the D. cardini group.




Similar content being viewed by others
References
Bächli G (2007) Taxodros database 2006/08: The database on taxonomy of Drosophilidae. URL: http://www.taxodros.unizh.ch/. Last accessed in April 2007
Brisson JA, Wilder J, Hollocher H (2006) Phylogenetic analysis of the cardini group of Drosophila with respect to changes in pigmentation. Evolution 60:1228–1241
Capy P, Bazin C, Higuet D, Langin T (1997) Dynamics and evolution of transposable elements. Landes Bioscience, Texas
Cizeron G, Lemeunier F, Loevenbruck C, Brehm A, Biémont C (1998) Distribution of the retrotransposable element 412 in Drosophila species. Mol Biol Evol 15:1589–1599
Da Cunha AB, Brncic D, Salzano FM (1953) A comparative study of chromosomal polymorphism in certain South American species of Drosophila. Heredity 2:193–202
Daniels SB, Chovnick A, Boussy IA (1990) Distribution of hobo transposable elements in the genus Drosophila. Mol Biol Evol 7:589–606
De Almeida LM, Carareto CMA (2004) Identification of two subfamilies of micropia transposable element in species of the repleta group of Drosophila. Genetica 121:155–164
De Almeida LM, Carareto CMA (2006) Sequence heterogeneity and phylogenetic relationships between the copia retrotransposon in Drosophila species of the repleta and melanogaster groups. Genet Sel Evol 38:535–550
De Almeida LM, Castro JP, Carareto CMA (2001) Micropia transposable element occurrence in Drosophila species of the saltans group. DIS 84:114–117
De Toni DC, Herédia FO, Valente VLS (2001) Chromosomal variability of Drosophila polymorpha populations from Atlantic Forest remnants of continental and insular environments in the State of Santa Catarina, Brazil. Caryologia 4:329–337
Felsenstein J (1985) Confidence limits on phylogenies: an approaching using bootstrap. Evolution 39:783–791
Hasegawa M, Kishino H, Yano T (1985) Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22:160–174
Heath BD, Butcher RD, Whitfield WG, Hubbard SF (1999) Horizontal transfer of Wolbachia between phylogenetically distant insect species by a naturally occurring mechanism. Curr Biol 9:313–316
Heed WB (1962) Genetic characteristics of island populations. Univ Texas Publ Stud Genet 6205:173–206
Heed WB (1963) Density and distribution of Drosophila polymorpha and its color alleles in south America. Evolution 17:502–518
Heed WB, Blake PR (1963) A new color allele at the e locus of Drosophila polymorpha from northern south America. Genetics 48:217–234
Heed WB, Krishnamurthy ND (1959) Genetic studies on the cardini group of Drosophila in the West Indies. Univ Texas Publs 5914:155–179
Heed WB, Russel JS (1971) Phylogeny and population structure in island and continental species of the cardini group of Drosophila studied by inversion analysis. Univ Texas Publs 7103:91–130
Hendy MD, Penny D (1982) Branch and bound algorithms to determine minimal evolution trees. Math Biosci 59:277–290
Herédia FO, Loreto ELS, Valente VLS (2004) Complex evolution of gypsy in drosophilid species. Mol Biol Evol 21:1–12
Hollocher H, Hatcher JL, Dyreson EG (2000a) Evolution of abdominal pigmentation differences across species in the Drosophila dunni subgroup. Evolution 54:2046–2056
Hollocher H, Hatcher JL, Dyreson EG (2000b) Genetic and developmental analysis of abdominal pigmentation differences across species in the Drosophila dunni subgroup. Evolution 54:2057–2071
Hoogland C, Vieira C, Biémont C (1997) Chromosomal distribution of the 412 retrotransposon in natural populations of Drosophila simulans. Heredity 79:128–134
Houck MA, Clark JB, Peterson HR, Kidwell MG (1991) Possible horizontal transfer of Drosophila genes by the mite Proctolaelaps regalis. Science 253:1125–1128
Huijser P, Kirchhoff C, Lankenau D-H, Hennig W (1988) Retrotransposon-like sequences are expressed in Y chromosomal lampbrush loops of Drosophila hydei. J Mol Biol 203:689–697
Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with ClustalX. Trends Biochem Sci 23:403–405
Jordan IK, Maayunina LV, McDonald JF (1999) Evidence for the recent horizontal transfer of long terminal repeat retrotransposon. PNAS 22:12621–12625
Kumar S, Tamura K, Nei M (2004) MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform 5:150–163
Lankenau D-H (1993) The retrotransposon family micropia in Drosophila species. In: McDonald J (eds) Transposable elements and evolution. Kluwer Publishers, Amsterdam pp 232–241
Lankenau D-H, Huijser P, Hennig W (1989) Characterization of the long terminal repeats of micropia elements microdissected from the Y-chromosomal lampbrush loops “threads” of Drosophila hydei. J Mol Biol 209:493–497
Lankenau D-H, Huijser P, Jansen E, Miedema K, Hennig W (1988) micropia: a retrotransposon of Drosophila combining structural features of DNA viruses, retroviruses and non-viral transposable elements. J Mol Biol 2:233–246
Lankenau D-H, Huijser P, Jansen E, Miedema K, Hennig W (1990) DNA sequence comparison of micropia transposable elements from Drosophila hydei and Drosophila melanogaster. Chromosoma 99:111–117
Lankenau S, Corces GV, Lankenau D-H (1994) The Drosophila micropia retrotransposon encodes a testis-specific antisense RNA complementary to reverse transcriptase. Mol Biol Evol 17:1542–1557
Loreto ELS, Basso da Silva L, Zaha A, Valente VLS (1998) Distribution of transposable elements in Neotropical species of Drosophila. Genetica 101:153–165
Ludwig A, Loreto ELS (2006) Evolutionary pattern of the gtwin retrotransposon in the Drosophila melanogaster subgroup. Genetica 130:161–168
Malogolowkin C (1953) Sobre a genitália dos drosofilídeos. IV. A genitália masculina no subgênero Drosophila (Diptera, Drosophilidae). Rev Bras Biol 13:245–264
Mejlumian L, Pelisson A, Bucheton A, Terzian C (2002) Comparative and functional studies of Drosophila species invasion by the gypsy endogenous retrovirus. Genetics 160:201–209
Montchamp-Moreau C, Ronsseray M, Jacques M, Lehmann M, Anxolabéhère D (1993) Distribution and conservation of sequences homologous to the 1731 retrotransposon in Drosophila. Mol Biol Evol 10:791–803
Napp M, Cordeiro AR (1981) Interspecific relationships in the cardini group of Drosophila studied by electrophoresis. Revta Bras Genet 4:537–547
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Nicholas KB, Nicholas HB (1997) GeneDoc: a tool for editing and annotations of multiple sequence alignements. Distributed by the author. http://www.psc.edu/biomed/genedoc
Posada C, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Robe LJ, Valente VLS, Budnik M, Loreto ELS (2005) Molecular phylogeny of the subgenus Drosophila (Diptera, Drosophilidae) with an emphasis on Neotropical species and groups: A nuclear versus mitochondrial gene approach. Mol Phylogenet Evol 3:623–640
Rohde C, Valente VLS (1996) Cytological maps and chromosomal polymorphism of Drosophila polymorpha and Drosophila cardinoides. Braz J Genet 19:27–32
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sassi AK, Herédia FO, Loreto ELS, Valente VLS, Rohde C (2005) Transposable elements P and gypsy in natural populations of Drosophila willistoni. Genet Mol Biol 28:734–739
Sharp PM, Li W-H (1989) On the rate of DNA sequence evolution in Drosophila. J Mol Evol 28:398–402
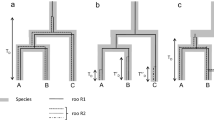
Silva JC, Loreto ELS, Clark JB (2004) Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol 6:57–72
Stacey SN, Lansman RA, Brock HW Grigliatti TA (1986) Distribution and conservation of mobile elements in the genus Drosophila. Mol Biol Evol 3:522–534
Staden R (1996) The Staden sequence analysis package. Mol Biotechnol 5:233–241
Swofford DL (2003) PAUP*. Phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer Associates, Sunderland, Massachusetts
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C biases. Mol Biol Evol 9:678–687
Throckmorton LH (1975) The phylogeny, ecology and geography of Drosophila. In: King RC (ed) Handbook of genetics. Plenum, New York, pp 421–469
Val FC, Vilela CR, Marques MD (1981) Drosophilidae of the Neotropical region. In: Ashburner M, Carson HL, Thompson JN Jr (eds) The genetics and biology of Drosophila. 3a. Academic Press, London, pp 123–168
Vieira C, Lepetit D, Dumont S, Biémont C (1999) Wake up of transposable elements following Drosophila simulans worldwide colonization. Mol Biol Evol 16:1251–1255
Vilela CR, da Silva AFG, Sene FM (2002) Preliminary data on the geographical distribution of Drosophila species within morphoclimatic domains of Brazil. Rev Bras Entomol 2:139–148
Wang D, Marsh JL, Ayala FJ (1996) Evolutionary changes in the expression pattern of a developmentally essential gene in three Drosophila species. Proc Natl Acad Sci USA 93:7103–7107
Wilder JA, Hollocher H (2001) Mobile elements and the genesis of microsatellites in dipterans. Mol Biol Evol 18:384–392
Acknowledgements
This work was supported by grants from CAPES, CNPq, and FAPERGS research funding. The authors thank MSc Adriana Ludwig for statistical support and discussion, Daniel F. Dombrosky for valuable help and the members of the Drosophila Laboratories from Universidade Federal do Rio Grande do Sul, Universidade Federal de Santa Catarina and Universidade Federal de Santa Maria. We thank also the two anonymous reviewers’ valuable suggestions, Dr. Claudia M. A. Carareto and Dr. Dirk-Henner Lankenau for kindly providing the dhMiF2 plasmid, and Félix Nonnemacher for English revision.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Cordeiro, J., Robe, L.J., Loreto, É.L.S. et al. The LTR retrotransposon micropia in the cardini group of Drosophila (Diptera: Drosophilidae): a possible case of horizontal transfer. Genetica 134, 335–344 (2008). https://doi.org/10.1007/s10709-008-9241-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-008-9241-2