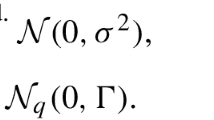Abstract
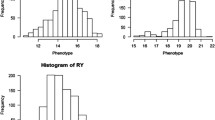
The maximum likelihood method of QTL mapping assumes that the phenotypic values of a quantitative trait follow a normal distribution. If the assumption is violated, some forms of transformation should be taken to make the assumption approximately true. The Box–Cox transformation is a general transformation method which can be applied to many different types of data. The flexibility of the Box–Cox transformation is due to a variable, called transformation factor, appearing in the Box–Cox formula. We developed a maximum likelihood method that treats the transformation factor as an unknown parameter, which is estimated from the data simultaneously along with the QTL parameters. The method makes an objective choice of data transformation and thus can be applied to QTL analysis for many different types of data. Simulation studies show that (1) Box–Cox transformation can substantially increase the power of QTL detection; (2) Box–Cox transformation can replace some specialized transformation methods that are commonly used in QTL mapping; and (3) applying the Box–Cox transformation to data already normally distributed does not harm the result.
Similar content being viewed by others
References
G.E.P. Box D.R. Cox (1964) ArticleTitleAn analysis of transformations J. Roy. Stat. Soc. B 26 211–252
K.W. Broman (2003) ArticleTitleQTL mapping in the case of a spike in the phenotype distribution Genetics 163 1169–1175 Occurrence Handle12663553
G.A. Churchill R.W. Doerge (1994) ArticleTitleEmpirical threshold values for quantitative trait mapping Genetics 138 963–971 Occurrence Handle7851788 Occurrence Handle1:STN:280:DyaK2M7lsVOiug%3D%3D
W. Coppieters A. Kvasz F. Farnir J.J. Arranz B. Grisart M. Mackinnon M. Georges (1998) ArticleTitleA rank-based nonparametric method for mapping quantitative trait loci in outbred half-sib pedigrees: Application to milk production in a granddaughter design Genetics 149 1547–1555 Occurrence Handle9649541 Occurrence Handle1:STN:280:DyaK1czhsFWitw%3D%3D
A.P. Dempster N.M. Laird D.B. Rubin (1977) ArticleTitleMaximum likelihood from incomplete data via EM algorithm J. Roy. Stat. Soc. Ser. B 39 1–38
R.C. Elston (1984) ArticleTitleThe genetic analysis of quantitative trait differences between two homozygous lines Genetics 108 733–744 Occurrence Handle6500262 Occurrence Handle1:STN:280:DyaL2M%2Flt1CktQ%3D%3D
Gray, G., 1992. Bias in misspecified mixtures, in Institute of Statistics Series 2227, North Carolina State University
R.G. Gutierrez R.J. Carroll N. Wang G.-H. Lee B.H. Taylor (1995) ArticleTitleAnalysis of tomato root initiation using a normal mixture distribution Biometrics 51 1461–1468 Occurrence Handle8589233 Occurrence Handle1:STN:280:DyaK287jsFChtA%3D%3D Occurrence Handle10.2307/2533276
C.A. Hackett (1997) ArticleTitleModel diagnostics for fitting QTL models to trait and marker data by interval mapping Heredity 79 319–328 Occurrence Handle10.1038/sj.hdy.6882150
C.A. Hackett J.I. Weller (1995) ArticleTitleGenetic mapping of quantitative trait loci for traits with ordinal distributions Biometrics 51 1252–1263 Occurrence Handle8589221 Occurrence Handle1:STN:280:DyaK287jsFCitw%3D%3D Occurrence Handle10.2307/2533257
C.S. Haley S.A. Knott J.M. Elsen (1994) ArticleTitleMapping quantitative trait loci in crosses between outbred lines using least squares Genetics 136 1195–1207 Occurrence Handle8005424 Occurrence Handle1:CAS:528:DyaK2MXpsFCr
R.C. Jansen (1992) ArticleTitleA general mixture model for mapping quantitative trait loci by using molecular markers Theor. Appl. Genet. 85 252–260 Occurrence Handle1:CAS:528:DyaK3sXktVantLs%3D Occurrence Handle10.1007/BF00222867
S.A. Knott C.S. Haley (1992) ArticleTitleAspects of maximum likelihood methods for the mapping of quantitative trait loci in line crosses Genet. Res. 60 139–151 Occurrence Handle10.1017/S0016672300030822
Y. Mao S. Xu (2004) ArticleTitleMapping QTLs for traits measured as percentages Genet. Res. 83 159–168 Occurrence Handle15462409 Occurrence Handle10.1017/S0016672304006834 Occurrence Handle1:CAS:528:DC%2BD2cXlsFyqtrk%3D
O. Martinez R.N. Curnow (1992) ArticleTitleEstimating the locations and the sizes of the effects of quantitative trait loci using flanking markers Theor. Appl. Genet. 85 480–488 Occurrence Handle10.1007/BF00222330
H. Muranty B. Goffinet (1997) ArticleTitleSelective genotyping for location and estimation of the effect of a quantitative trait locus Biometrics 53 629–643 Occurrence Handle10.2307/2533963
S.Q. Rao S.Z. Xu (1998) ArticleTitleMapping quantitative trait loci for ordered categorical traits in four-way crosses Heredity 81 214–224 Occurrence Handle9750263 Occurrence Handle10.1046/j.1365-2540.1998.00378.x
A. Rebaï (1997) ArticleTitleComparison of methods for regression interval mapping in QTL analysis with non-normal traits Genet. Res. 69 69–74 Occurrence Handle10.1017/S0016672396002558
R.R. Sokal F.J. Rohlf (1995) Biometry: The Principles and Practice of Statistics in Biological Research W.H. Freeman and Company New York
S.Z. Xu W.R. Atchley (1996) ArticleTitleMapping quantitative trait loci for complex binary diseases using line crosses Genetics 143 1417–1424 Occurrence Handle8807312 Occurrence Handle1:STN:280:DyaK28vgtV2gtA%3D%3D
S. Xu N. Yi D. Burke A. Galecki R.A. Miller (2003) ArticleTitleAn EM algorithm for mapping binary disease loci: Application to fibrosarcoma in a four-way cross mouse family Genet. Res. 82 127–138 Occurrence Handle14768897 Occurrence Handle1:CAS:528:DC%2BD3sXovVKmsLg%3D Occurrence Handle10.1017/S0016672303006414
Zar, J.H., 1996. Biostatistical Analysis, 3rd edn. Prentice Hall
Zou, F., 2001. Efficient and robust statistical methodologies for quantitative trait loci analysis. PhD Thesis, Statistics, University of Wisconsin, Madison
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, R., Yi, N. & Xu, S. Box–Cox transformation for QTL mapping. Genetica 128, 133–143 (2006). https://doi.org/10.1007/s10709-005-5577-z
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10709-005-5577-z