Abstract
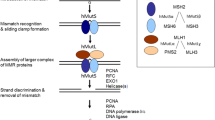
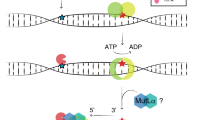
Missense mutations of the DNA mismatch repair gene MLH1 are found in a significant fraction of patients with Lynch syndrome (hereditary nonpolyposis colorectal cancer, HNPCC) and their pathogenicity often remains unclear. We report here all 88 MLH1 missense variants identified in families from the German HNPCC consortium with clinical details of these patients/families. We investigated 23 MLH1 missense variants by two functional in vivo assays in yeast; seven map to the ATPase and 16 to the protein interaction domain. In the yeast-2-hybrid (Y2H) assay three variants in the ATPase and twelve variants in the interaction domain showed no or a reduced interaction with PMS2; seven showed a normal and one a significantly higher interaction. Using the Lys2A 14 reporter system to study the dominant negative mutator effect (DNE), 16 variants showed no or a low mutator effect, suggesting that these are nonfunctional, three were intermediate and four wild type in this assay. The DNE and Y2H results were concordant for all variants in the interaction domain, whereas slightly divergent results were obtained for variants in the ATPase domain. Analysis of the stability of the missense proteins in yeast and human embryonic kidney cells (293T) revealed a very low expression for seven of the variants in yeast and for nine in human cells. In total 15 variants were classified as deleterious, five were classified as variants of unclassified significance (VUS) and three were basically normal in the functional assays, P603R, K618R, Q689R, suggesting that these are neutral.



Similar content being viewed by others
References
Guarne A, Ramon-Maiques S, Wolff EM et al (2004) Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. EMBO J 23(21):4134–4145
Kunkel TA, Erie DA (2005) DNA mismatch repair. Annu Rev Biochem 74:681–710
Matson SW, Robertson AB (2006) The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic Acids Res 34(15):4089–4097
Modrich P (2006) Mechanisms in eukaryotic mismatch repair. J Biol Chem 281(41):30305–30309
Guerrette S, Acharya S, Fishel R (1999) The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem 274(10):6336–6341
Kondo E, Suzuki H, Horii A, Fukushige S (2003) A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res 63(12):3302–3308
Nystrom-Lahti M, Perrera C, Raschle M et al (2002) Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer 33(2):160–167
Fishel R (2001) The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res 61(20):7369–7374
Harfe BD, Jinks-Robertson S (2000) DNA mismatch repair and genetic instability. Annu Rev Genet 34:359–399
Jiricny J (2000) Mediating mismatch repair. Nat Genet 24(1):6–8
Plotz G, Welsch C, Giron-Monzon L et al (2006) Mutations in the MutSalpha interaction interface of MLH1 can abolish DNA mismatch repair. Nucleic Acids Res 34(22):6574–6586
Fedier A, Fink D (2004) Mutations in DNA mismatch repair genes: implications for DNA damage signaling and drug sensitivity (review). Int J Oncol 24(4):1039–1047
de la Chapelle A (2004) Genetic predisposition to colorectal cancer. Nat Rev Cancer 4(10):769–780
Fields S, Song O (1989) A novel genetic system to detect protein-protein interactions. Nature 340(6230):245–246
Shcherbakova PV, Kunkel TA (1999) Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol 19(4):3177–3183
Shimodaira H, Filosi N, Shibata H et al (1998) Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet 19(4):384–389
Trojan J, Zeuzem S, Randolph A et al (2002) Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology 122(1):211–219
Raevaara TE, Korhonen MK, Lohi H et al (2005) Functional significance and clinical phenotype of nontruncating mismatch repair variants of MLH1. Gastroenterology 129(2):537–549
Raevaara TE, Gerdes AM, Lonnqvist KE et al (2004) HNPCC mutation MLH1 P648S makes the functional protein unstable, and homozygosity predisposes to mild neurofibromatosis type 1. Genes Chromosom Cancer 40(3):261–265
Stone EA, Sidow A (2005) Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res 15(7):978–986
Kosinski J, Hinrichsen I, Bujnicki JM, Friedhoff P, Plotz G (2010) Identification of Lynch syndrome mutations in the MLH1-PMS2 interface that disturb dimerization and mismatch repair. Hum Mutat 31(8):975–982
Takahashi M, Shimodaira H, Andreutti-Zaugg C, Iggo R, Kolodner RD, Ishioka C (2007) Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res 67(10):4595–4604
Kondo E, Horii A, Fukushige S (1999) The human PMS2L proteins do not interact with hMLH1, a major DNA mismatch repair protein. J Biochem 125(4):818–825
Clark AB, Cook ME, Tran HT, Gordenin DA, Resnick MA, Kunkel TA (1999) Functional analysis of human MutSalpha and MutSbeta complexes in yeast. Nucleic Acids Res 27(3):736–742
Mielke C, Tummler M, Schubeler D, von Hoegen I, Hauser H (2000) Stabilized, long-term expression of heterodimeric proteins from tricistronic mRNA. Gene 254(1–2):1–8
Agatep R, Kirkpatrick RD, Parchaliuk DL, Woods RA, Gietz RD (1998) Transformation of Saccharomyces cerevisiae by the lithium acetate/single-stranded carrier DNA/polyethylene glycol (LiAc/ss-DNA/PEG) protocol. Technical Tips Online (http://tto.trends.com)
Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA (1997) Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol 17(5):2859–2865
Lea DC (1949) The distribution of the numbers of mutants in bacterial populations. J Genet 49:264–285
Kondo E, Horii A, Fukushige S (2001) The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res 29(8):1695–1702
Wanat JJ, Singh N, Alani E (2007) The effect of genetic background on the function of Saccharomyces cerevisiae mlh1 alleles that correspond to HNPCC missense mutations. Hum Mol Genet 16(4):445–452
Chao EC, Velasquez JL, Witherspoon MS et al (2008) Accurate classification of MLH1/MSH2 missense variants with multivariate analysis of protein polymorphisms-mismatch repair (MAPP-MMR). Hum Mutat 29(6):852–860
Muller-Koch Y, Kopp R, Lohse P et al (2001) Sixteen rare sequence variants of the hMLH1 and hMSH2 genes found in a cohort of 254 suspected HNPCC (hereditary non-polyposis colorectal cancer) patients: mutations or polymorphisms? Eur J Med Res 6(11):473–482
Pang Q, Prolla TA, Liskay RM (1997) Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol 17(8):4465–4473
Prolla TA, Pang Q, Alani E, Kolodner RD, Liskay RM (1994) MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science 265(5175):1091–1093
Acknowledgments
This work was supported by grants from the German Cancer Aid (Deutsche Krebshilfe e.V. Bonn, project nos 70-2397, 108132 and 108628) and a multicentre grant from the German Cancer Aid (project nos. 70-2371, 106244 and 107318).
Author information
Authors and Affiliations
Consortia
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hardt, K., Heick, S.B., Betz, B. et al. Missense variants in hMLH1 identified in patients from the German HNPCC consortium and functional studies. Familial Cancer 10, 273–284 (2011). https://doi.org/10.1007/s10689-011-9431-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-011-9431-4