Abstract
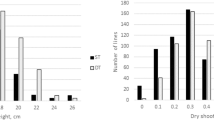
Soil salinity negatively affects growth and development as well as yield and fiber quality of cotton. The identification of quantitative trait loci (QTLs) for traits related to salt tolerance could facilitate the development of cotton cultivars with salt tolerance. The objective of this study was to map QTLs for salt tolerance in an F2:3 population derived from an interspecific cross between an upland cotton, CRI-12 (G09091801-2), of upland cotton (Gossypium hirsutum) and an accession, AD3-00 (P0601211), of wild cotton Gossypium tomentosum. 1295 simple sequence repeat markers, which amplified 1342 loci, distributed on 26 chromosomes and covered 3328.24 cM with an average inter-marker distance of 3.0 cM, were utilized for molecular genotyping. Salt tolerance was evaluated in a hydroponic at a young seedling stage for 2 weeks at 150 mM NaCl concentration in three environments. Mapping of QTLs related to salt tolerance was carried out on 7 traits by composite interval mapping using Windows QTL Cartographer 2.5. Eleven consistent QTLs were detected on 8 chromosomes (9, 11, 15, 16, 21, 23, 24 and 26) in at least two environments. qRL-Chr16-1 for RL was a major QTL explaining the Phenotypic variance of 11.97 and 18.44 % in two environments. Of the 11 QTLs, 10 were located on the D subgenome, indicating that genes responsible for salt tolerance in the allotetraploid cotton AD genome were mainly derived from the D subgenome. The information derived from these studies may be useful in facilitating breeding of salt tolerant cotton lines.

Similar content being viewed by others
References
Ashraf M, Foolad MR (2013) Crop breeding for salt tolerance in the era of molecular markers and marker-assisted selection. Plant Breed 132:10–20. doi:10.1111/pbr.12000
Azhar FM, McNeilly T (1988) The genetic basis of variation for salt tolerance in Sorghum bicolor (L.) Moench seedlings. Plant Breed 101:114–121. doi:10.1111/j.1439-0523.1988.tb00275.x
Bhatti MA, Azhar FM (2002) Salt tolerance of nine Gossypium hirsutum L. varieties to NaCl salinity at early stage of plant development. Int J Agric Biol 4:544–546
Chee P, Draye X, Jiang C-X et al (2005) Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor Appl Genet 111:757–763. doi:10.1007/s00122-005-2063-z
Cho SK, Ryu MY, Song C et al (2008) Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20:1899–1914
DeJoode DR, Wendel JF (1992) Genetic diversity and origin of the hawaiian islands cotton, Gossypium tomentosum. Am J Bot 79:1311–1319
DeVicente MC, Tanksley SD (1993) QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134:585–596
Epstein E, Norlyn JD (1977) Seawater-based crop production: a feasibility study. Science 197:249–251. doi:10.1126/science.197.4300.249
Foolad MR, Jones RA (1993) Mapping salt-tolerance genes in tomato (Lycopersicon esculentum) using trait-based marker analysis. Theor Appl Genet Int J Plant Breed Res 87:184–192. doi:10.1007/BF00223763
Fryxell PA (1979) The natural history of cotton tribe (Malvaceae, Tribe Gossypieae). Texas A&M University Press, College station
Genc Y, MCdonald GK, Tester M (2007) Reassessment of tissue Na + concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ 30:1486–1498. doi:10.1111/j.1365-3040.2007.01726.x
Genc Y, Oldach K, Verbyla A et al (2010) Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. TAG Theor Appl Genet 121:877–894. doi:10.1007/s00122-010-1357-y
Gorham J, Bridges J (1995) Effects of calcium on growth and leaf ion concentrations of Gossypium hirsutum grown in saline hydroponic culture. Plant Soil 176:219–227
Gossett DR, Millhollon EP, Caldwell WD, Mundy S (1991) Isozyme variation among salt tolerant and salt sensitive varieties of cotton. Proc Beltwide Cotton Res Conf Natl Cotton Council, Memphis, pp 556–559
Grover CE, Zhu X, Grupp KK et al (2015) Molecular confirmation of species status for the allopolyploid cotton species, Gossypium ekmanianum Wittmack. Genet Resour Crop Evol 62:103–114. doi:10.1007/s10722-014-0138-x
Hallauer AR, Carena MJ, Miranda Filho JB (1990) Quantitative genetics in maize breeding. Crop Res 23:78–79. doi:10.1016/0378-4290(90)90102-H
Hanif M, Noor E, Murtaza N et al (2008) Assessment of variability for salt tolerance at seedling stage in Gossypium hirsutum L. J Food Agric Environ 6:134–138
Higbie SM, Wang F, Stewart JM et al (2010) Physiological response to salt (NaCl) stress in selected cultivated tetraploid cottons. Int J Agron 2010:1–12. doi:10.1155/2010/643475
Hoagland DR and Arnon DI (1950) The water-culture method for growing plants without soil. California Experiment Station Circular No. 347. The College of Agriculture, University of California, Berkeley
Hulse-Kemp AM, Ashrafi H, Zheng X et al (2014) Development and bin mapping of gene-associated interspecific SNPs for cotton (Gossypium hirsutum L.) introgression breeding efforts. BMC Genom 15:945. doi:10.1186/1471-2164-15-945
Ibrahim M, Akhtar J, Younis M Riaz, Anwar-ul-Haq MA, Tahir M (2007) Selection of cotton (Gossypium hirsutum L.) genotypes against NaCl stress. Soil & Env 26:59–63
Jiang C-X, Chee PW, Draye X et al (2000) Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (Cotton). Evolution (N Y) 54:798–814
Joshi R, Ramanarao MV, Lee S et al (2014) Ectopic expression of ADP ribosylation factor 1 (SaARF1) from smooth cordgrass (Spartina alterniflora Loisel) confers drought and salt tolerance in transgenic rice and Arabidopsis. Plant Cell Tissue Organ Cult 117:17–30. doi:10.1007/s11240-013-0416-x
Khorsandi F, Anagholi A (2009) Reproductive compensation of cotton after salt stress relief at different growth stages. J Agron Crop Sci 195:278–283. doi:10.1111/j.1439-037X.2009.00370.x
Kosambi DD (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247
Leidi EO (1994) Genotypic variation of cotton in response to stress by NaCl or PEG. In: Peeters MC (ed) Cotton biotechnology., REUR technical series 32FAO, Rome, pp 67–73
Li F, Fan G, Lu C et al (2015) Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol 33:524–530. doi:10.1038/nbt.3208
Liang Q, Hu C, Hua H et al (2013) Construction of a linkage map and QTL mapping for fiber quality traits in upland cotton (Gossypium hirsutum L.). Chin Sci Bull 58:3233–3243. doi:10.1007/s11434-013-5807-1
Liem ASN, Hendriks A, Kraal H, Loenen M (1985) Effects of de-icing salt on roadside grasses and herbs. Plant Soil 84:299–310
Liu S, Saha S, Burr B, Cantrell RG (2000) Chromosomal assigment of microsatellite loci in cotton. J Hered 91(4):326–332
Maas EV (1990) Crop salt tolerance. Chapter 13, In: Tanji KK (ed.). Agricultural salinity assessment and management. ASCE Manual and Reports on Engineering Practice No. 71. American Society of Civil Engineers, pp 262–304
MacCaughey V (1917) Vegetation of Hawaiian lava flows. Bot Gaz 64:386–420
Mano Y, Takeda K (1997) Mapping quantitative trait loci for salt tolerance at germination and the seedling stage in barley (Hordeum vulgare L.). Euphytica 94:263–272. doi:10.1023/A:1002968207362
Mehboob-ur-Rahman, Zafar Y, Paterson AH (2009) Gossypium DNA markers: types, numbers, and uses. In: Paterson AH (ed) Genetics and genomics of cotton. Plant genetics and genomics: Crops and Models, vol 3. Springer, pp 101–139
Mehetre SS, Patil SC, Pawar SV, Pardedhi SU, Shinde GC, Aher AR (2004) Ovulo embryo cultured hybrid between amphidiploids (Gossypium arboretum & Gossypium anomalum) and Gossypium hirsutum. Curr Sci 87:286–289
Mo H, Pua E-C (2002) Up-regulation of arginine decarboxylase gene expression and accumulation of polyamines in mustard (Brassica juncea) in response to stress. Physiol Plant 114:439–449
Munns R (1980) Mechanisms of salt tolerance in nonhalophytes. Plant Physiol 31:149–190
Munns R (2007) Utilizing genetic resources to enhance productivity of salt-prone land. CAB Rev 009:1–11
Nadarajan NGM (2005) Quantitative genetics and biometrical techniques in plant breeding. Kalyani Publication, New Delhi
Nguyen TB, Giband M, Brottier P et al (2004) Wide coverage of the tetraploid cotton genome using newly developed microsatellite markers. Theor Appl Genet 109:167–175. doi:10.1007/s00122-004-1612-1
Park YH, Alabady MS, Ulloa M et al (2005) Genetic mapping of new cotton fiber loci using EST-derived microsatellites in an interspecific recombinant inbred line cotton population. Mol Genet Genomics 274:428–441
Paterson AH, DeVerna JW, Lanini B, Tanksley SD (1990) Fine mapping of quantitative trait loci using selected overlapping recombinant chromosomes, in an interspecies cross of tomato. Genetics 124:735–742
Paterson AH, Lin Y-R, Li Z, Schertz KF, Doebley JF et al (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269:1714–1718
Ponnamperuma FN (1984) Role of cultivar tolerance in increasing rice production on saline lands. In: Staples RC, Toenniessen GH (eds) Salinity tolerance in plants. Wiley, New York, pp 255–272
Qadir M, Shams M (1997) Some agronomic and physiological aspects of salt tolerance in cotton (Gossypium hirsutum L.). J Agron Crop Sci 179:101–106. doi:10.1111/j.1439-037X.1997.tb00504.x
Quesada V, García-Martínez S, Piqueras P, Ponce MR, Micol JL (2002) Genetic architecture of NaCl tolerance in Arabidopsis thaliana. Plant Physiol 130:951–963
Rengasamy P (2006) World salinization with emphasis on Australia. J Exp Bot 57:1017–1023. doi:10.1093/jxb/erj108
Riaz MK (2013) A step towards cotton genome assembly through construction of high density genetic map from interspecific cross of G. hirsutum × G. tomentosum. Dissertation Institute of Cotton Research, Chinese Academy of Agricultural Sciences
Rick CM, Smith PG (1953) Novel variation in tomato species hybrids. Am Nat 87:359–373
Rong J, Abbey C, Bowers JE et al (2004) A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166:389–417. doi:10.1534/genetics.166.1.389
Rungis D, Llewellyn D, Dennis ES, Lyon BR (2005) Simple sequence repeat (SSR) markers reveal low levels of polymorphism between cotton (Gossypium hirsutum L.) cultivars. Aust J Agric Res 56:301–307
Saba J, Moghaddam M, Ghassemi K, Nishabouri MR (2001) Genetic properties of drought resistance indices. J Agric Sci Technol 3:43–49
Saeed M, Wangzhen G, Tianzhen Z (2014) Association mapping for salinity tolerance in cotton (Gossypium hirsutum L.) germplasm from US and diverse regions of China. Aust J Crop Sci 8:338–346
Stephens SG (1964) Native Hawaiian cotton (Gossypium tomentosum Nutt.). Pac Sci 18:385–398
Stuber CW, Edwards MD, Wendel JF (1987) Molecular marker facilitated investigations of quantitative trait loci in maize. II. Factors influencing yield and its component traits. Crop Sci 27:639–648
Takehisa H, Shimodate T, Fukuta Y et al (2004) Identification of quantitative trait loci for plant growth of rice in paddy field flooded with salt water. Field Crop Res 89:85–95. doi:10.1016/j.fcr.2004.01.026
Tiwari RS, Stewart JM (2008) Effect of Salt on Several Genotypes of Gossypium. AES Res Ser 573:34–36
Tiwari RS, Picchioni GA, Steiner RL et al (2013) Genetic variation in salt tolerance at the seedling stage in an interspecific backcross inbred line population of cultivated tetraploid cotton. Euphytica 194:1–11. doi:10.1007/s10681-013-0927-x
Van Ooijen JW (2006) JoinMap ® 4; Software for the calculation of genetic linkage maps in experimental populations, version Wageningen. Kyazma B.V, Wageningen
Vega U, Frey KJ (1980) Transgressive segregation in inter and intraspecific crosses of barley. Euphytica 29:585–594
Villalta I, Reina-Sánchez A, Bolarín MC et al (2008) Genetic analysis of Na + and K + concentrations in leaf and stem as physiological components of salt tolerance in Tomato. Theor Appl Genet 116:869–880. doi:10.1007/s00122-008-0720-8
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. doi:10.1093/jhered/93.1.77
Wang K, Song X, Han Z et al (2006) Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet 113:73–80. doi:10.1007/s00122-006-0273-7
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics. North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wendel JF, Brubaker C, Alvarez I, Cronn R, Stewart JM (2009) Evolution and natural history of the cotton Genus. In: Paterson AH (ed) Genetics and genomics of cotton, vol 3. Springer, New York, pp p3–p22
Young ND, Tanksley SD (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet 77:95–101. doi:10.1007/BF00292322
Youngner VB, Lunt OR (1967) Salinity effects on roots and tops of bermuda grass. Grass Forage Sci 22:257–259. doi:10.1111/j.1365-2494.1967.tb00536.x
Zhang L, W Ye, J Wang, B Fan (2010) Studies of salinity-tolerance with SSR markers on G. hirsutum L. Cott Sci 22:175–180. http://www.journal.cricaas.com.cn/en/2010/02/cs100214.htm
Zhang T, Hu Y, Jiang W et al (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537. doi:10.1038/nbt.3207
Zhu J-K (2011) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. doi:10.1146/annurev.arplant.53.091401.143329
Acknowledgments
This program was financially sponsored by the National Natural Science Foundation of China(31530053; 31401424).
Author contributions
KBW & FL designed the experiments. OG and JYZ conceived the experiments and analyzed the results having equal contribution. OG & XXW carried out most computational analyses. JYZ and OG carried out the experiments. ZLZ, MKR, XYC, CYW and YHW participated in part of QTL analysis. XYL and HW participated in part of trait collection. OG and JYZ drafted the manuscript. XXW and KBW revised the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
George Oluoch and Juyun Zheng have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Oluoch, G., Zheng, J., Wang, X. et al. QTL mapping for salt tolerance at seedling stage in the interspecific cross of Gossypium tomentosum with Gossypium hirsutum . Euphytica 209, 223–235 (2016). https://doi.org/10.1007/s10681-016-1674-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-016-1674-6