Abstract
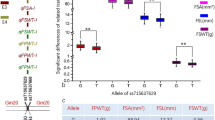
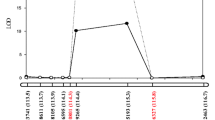
Glycine soja has the potential to improve upon the cultivated soybean (G. max) in protein content, resistance/tolerance to stresses, and other traits, but its small 100-seed weight (100SW) is usually undesired. To explore the 100SW QTL-allele system of G. soja, the recombinant inbred line population NJRINY with 286 lines was developed from a cross between G. max (NN86-4, 17.9 g) and G. soja (PI342618B, 1.1 g) and was tested over 4 years. The genetic linkage map was constructed with 181 PAV (presence/absence variation) and 42 SSR markers. The quantitative trait locus (QTL) mapping results showed that the wild genetic system of 100SW was composed of three groups of QTLs, including (1) 15 additive QTLs, which accounted for 55.1 % of the phenotypic variation (PV), with each locus contributing 1.5–8.1 % of the PV and all of the wild alleles having negative effects (−0.1 to −0.5 g); (2) 17 epistatic QTL pairs, which accounted for 19.0 % of the PV, with epistatic effects positive for parental type (0.1–0.2 g) and negative for recombinants and all of the QTLs in epistatic pairs having simultaneously additive effects except one pair (SW16); (3) a collection of unmapped minor QTLs that accounted for 22.9 % of the PV. Among the detected total additive and/or epistatic QTLs, 12 QTLs have been reported in the literature, but five of the QTLs and all of the epistatic pairs have not been reported before. The results can be used for marker-assisted breeding for 100SW.



Similar content being viewed by others
References
Broich SL, Palmer RG (1980) A cluster analysis of wild and domesticated soybean phenotypes. Euphytica 29:23–32
Carter TE Jr, Nelson RL, Sneller CH, Cui Z (2004) Genetic diversity in soybean. In: Boerma HR, Specht JE (eds) Soybeans: improvement, production, and uses, 3rd edn. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, pp 303–416
Concibido VC, La Vallee B, Mclaird P, Pineda N, Meyer J, Hummel L, Yang J, Wu K, Delannay X (2003) Introgression of a quantitative trait locus for yield from Glycine soja into commercial soybean cultivars. Theor Appl Genet 106:575–582
Cregan PB, Jarvik T, Bush AL, Shoemaker RC, Lark KG, Kahler AL, Kaya N, VanToai TT, Lohnes DG, Chung J (1999) An integrated genetic linkage map of the soybean genome. Crop Sci 39:1464–1490
Csanádi G, Vollmann J, Stift G, Lelley T (2001) Seed quality QTLs identified in a molecular map of early maturing soybean. Theor Appl Genet 103:912–919
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Feng F, Luo L, Li Y, Zhou L, Xu X, Wu J, Chen H, Chen L, Mei H (2005) Comparative analysis of polymorphism of InDel and SSR markers in rice. Mol Plant Breed 3:725–730
Hartwig EE (1973) Varietal development. Soybeans: improvement, production and uses. American Society of Agronomy, Madison
Hoeck JA, Fehr WR, Shoemaker RC, Welke GA, Johnson SL, Cianzio SR (2003) Molecular marker analysis of seed size in soybean. Crop Sci 43:68–74
Kato S, Sayama T, Fujii K, Yumoto S, Kono Y, Hwang T-Y, Kikuchi A, Takada Y, Tanaka Y, Shiraiwa T, Ishimoto M (2014) A major and stable QTL associated with seed weight in soybean across multiple environments and genetic backgrounds. Theor Appl Genet 127:1365–1374
Kim HK, Kim YC, Kim ST, Son BG, Choi YW, Kang JS, Park YH, Cho YS, Choi IS (2010) Analysis of quantitative trait loci (QTLs) for seed size and fatty acid composition using recombinant inbred lines in soybean. J Life Sci 20:1186–1192
Kim M, Hyten DL, Niblack TL, Diers BW (2011) Stacking resistance alleles from wild and domestic soybean sources improves soybean cyst nematode resistance. Crop Sci 51:934–943
Kim H, Xing G, Wang Y, Zhao T, Yu D, Yang S, Li Y, Chen S, Palmer R, Gai J (2014) Constitution of resistance to common cutworm in terms of antibiosis and antixenosis in soybean RIL populations. Euphytica 196:137–154
Lam HM, Xu X, Liu X, Chen W, Yang G, Wong FL, Li MW, He W, Qin N, Wang B, Li J, Jian M, Wang J, Shao G, Wang J, Sun SSM, Zhang G (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059
Li DD, Pfeiffer TW, Cornelius PL (2008) Soybean QTL for yield and yield components associated with Glycine soja alleles. Crop Sci 48:571–581
Li Y-h, Liu B, Reif JC, Liu Y-l, Li H-h, Chang R-z, Qiu L-ju (2014) Development of insertion and deletion markers based on biparental resequencing for fine mapping seed weight in soybean. Plant Gen. doi:10.3835/plantgenome2014.04.0014
Liu B, Fujita T, Yan ZH, Sakamoto S, Xu D, Abe J (2007) QTL mapping of domestication-related traits in soybean (Glycine max). Ann Bot 100(5):1027–1038
Liu Y-l, Li Y-h, Reif JC, Mette MF, Liu Z-x, Liu B, Zhang S-s, Yan L, Chang R-z, Qiu L-j (2013) Identification of quantitative trait loci underlying plant height and seed weight in soybean. Plant Gen. doi:10.3835/plantgenome2013.03.0006
Mahama AA, Lewers KS, Palmer RG (2002) Genetic linkage in soybean. Crop Sci 42:1459–1464
Maughan PJ, Maroof MAS, Buss GR (1996) Molecular-marker analysis of seed-weight: genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theor Appl Genet 93:574–579
Nyquist WE, Baker R (1991) Estimation of heritability and prediction of selection response in plant populations. Crit Rev Plant Sci 10:235–322
Ooijen JW, Voorrips R (2002) JoinMap: version 3.0: software for the calculation of genetic linkage maps. University and Research Center, Plant Research International, Wageningen
Orf JH, Chase K, Jarvik T, Mansur LM, Cregan PB, Adler FR, Lark KG (1999) Genetics of soybean agronomic traits: I. Comparison of three related recombinant inbred populations. Crop Sci 39:1652–1656
Panaud O, Chen X, McCouch SR (1996) Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet 252:597–607
SAS Institute Inc (2002) SAS/STAT software: statistical analysis software version 9.1. SAS Institute Inc, Cary, North Carolina
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Sebolt AM, Shoemaker RC, Diers BW (2000) Analysis of a quantitative trait locus allele from wild soybean that increases seed protein concentration in soybean. Crop Sci 40:1438–1444
Song QJ, Marek LF, Shoemaker RC, Lark KG, Concibido VC, Delannay X, Specht JE, Cregan PB (2004) A new integrated genetic linkage map of the soybean. Theor Appl Genet 109:122–128
Song QJ, Jia GF, Zhu YL, Grant D, Nelson RT, Hwang EY, Hyten DL, Cregan PB (2010) Abundance of SSR motifs and development of candidate polymorphic SSR markers (BARCSOYSSR_1.0) in soybean. Crop Sci 50:1950–1960
Su CF, Lu WG, Zhao TJ (2010) Verification and fine-mapping of QTLs conferring days to flowering in soybean using residual heterozygous lines. Chin Sci Bull 55:499–508
Sun Y-n, Pan J-b, Shi X-l, Du X-y, Wu Q, Qi Z-m, Jiang H-w, Xin D-w, Liu C-y, Hu G-h, Chen Q-s (2012) Multi-environment mapping and meta-analysis of 100-seed weight in soybean. Mol Biol Rep 39:9435–9443
Tuyen DD, Lal SK, Xu DH (2010) Identification of a major QTL allele from wild soybean (Glycine soja Sieb. & Zucc.) for increasing alkaline salt tolerance in soybean. Theor Appl Genet 121:229–236
Voorrips RE (2002) MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang D, Graef GL, Procopiuk AM, Diers BW (2004) Identification of putative QTL that underlie yield in interspecific soybean backcross populations. Theor Appl Genet 108:458–467
Wang W, He Q, Yang H, Xiang S, Zhao T, Gai J (2013) Development of a chromosome segment substitution line population with wild soybean (Glycine soja Sieb. et Zucc.) as donor parent. Euphytica 189:293–307
Wang W, He Q, Yang H, Xiang S, Xing G, Zhao T, Gai J (2014a) Identification of QTL/segments related to seed-quality traits in G. soja using chromosome segment substitution lines. Plant Genet Resour C 12:S65–S69
Wang Y, Lu J, Chen S, Shu L, Palmer RG, Xing G, Li Y, Yang S, Yu D, Zhao T, Gai J (2014b) Exploration of presence/absence variation and corresponding polymorphic markers in soybean genome. J Integr Plant Biol 56:1009–1019
Yan L, Li Y-H, Yang C-Y, Ren S-X, Chang R-Z, Zhang M-C, Qiu L-J (2014) Identification and validation of an over-dominant QTL controlling soybean seed weight using populations derived from Glycine max × Glycine soja. Plant Breed 133:632–637
Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536
Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X, Zhu J (2008) QTLNetwork: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723
Yang K, Moon J-K, Jeong N, Chun H-K, Kang S-T, Back K, Jeong S-C (2011) Novel major quantitative trait loci regulating the content of isoflavone in soybean seeds. Genes Genom 33:685–692
Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, Wu XL, Gai JY, Chen SY (2004) QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet 108:1131–1139
Acknowledgments
This research was supported by the National Key Basic Research Program of China (2011CB1093), National Hightech R&D Program of China (2011AA10A105, 2012AA101106), MOE 111 Project (B08025), MOE Program for Changjiang Scholars and Innovative Research Team in University (PCSIRT13073), MOA Public Profit Program (201203026-4), Jiangsu Higher Education PAPD Program, and Jiangsu JCIC-MCP program.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Wubin Wang and Xuliang Li have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wang, W., Li, X., Chen, S. et al. Using presence/absence variation markers to identify the QTL/allele system that confers the small seed trait in wild soybean (Glycine soja Sieb. & Zucc.). Euphytica 208, 101–111 (2016). https://doi.org/10.1007/s10681-015-1591-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-015-1591-0