Abstract
A previously unknown haplotype of the plant pathogen ‘Candidatus Liberibacter solanacearum’ (Lso) was found in cultivated carrots and parsnips in eastern Finland. That same haplotype was found in western Finland, over 300 km away, in the family Polygonaceae, the species Fallopia convolvulus (wild buckwheat) and Persicaria lapathifolia (pale persicaria) growing as weeds within carrot and parsnip fields. The infected plants, both apiaceous and polygonaceous, showed symptoms of foliar discolouration. This is the first report of Lso bacteria in plants of the family Polygonaceae. The finding that the polygonaceous plants infected with a previously unknown haplotype of Lso were growing among the apiaceous plants infected with Lso haplotype C suggests that these two haplotypes might be transmitted by different vectors. Phylogenetic analyses showed that the new haplotype, called haplotype H, is distinct from the previously characterized haplotypes and appears to have diverged early from their common ancestor. Multi-locus sequence analysis revealed four different sequence types (strains) within the haplotype H. These findings suggest that the haplotype H is likely to be endemic in northern Europe and that the genetic diversity within the Lso species is higher than previously assumed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
‘Candidatus Liberibacter solanacearum’ (Lso; Liefting et al. 2009) is a Gram negative plant-pathogenic bacterium transmitted by psyllids. When feeding on the phloem sap of the host plant, the psyllids harbouring Lso deliver the bacteria into the phloem sieve cells, where the bacteria may then multiply and colonize the phloem systemically. Because of the dependency on a vector in accessing a plant, the natural host range of Lso is determined by the host plant range of the vector. Different psyllid species feeding on different plants and occurring in different geographical regions have been found to harbour diverse haplotypes of Lso. Thus far, eight distinct haplotypes have been found and named A, B, C, D, E, F, G, and U. The haplotypes A, B and F are associated with zebra chip disease of potato in America - the haplotype A also in New Zealand - and are transmitted by the tomato/potato psyllid Bactericera cockerelli (Hansen et al. 2008; Nelson et al. 2011; Swisher Grimm and Garczynski 2019). The haplotypes C, D and E have been found in Europe, D and E also in North Africa and D in the eastern Mediterranean area. These haplotypes are associated with diseases in apiaceous plants, including carrot and celery (Hajri et al. 2017; Mawassi et al. 2018; Munyaneza et al. 2010; Nelson et al. 2013; Tahzima et al. 2014; Teresani et al. 2014). In northern Europe, Lso haplotype C is transmitted by the carrot psyllid Trioza apicalis (Nissinen et al. 2014), whereas in the Mediterranean region the haplotypes D and E are transmitted by the psyllid Bactericera trigonica (Antolinez et al. 2017; Mawassi et al. 2018). In addition to T. apicalis, Lso haplotype C was found in Trioza anthrisci and its host plant Anthriscus sylvestris. Haplotype U was found in Trioza urticae and its nettle host plant (Urtica dioica) in Finland (Haapalainen et al. 2018). Recently, another haplotype of Lso was discovered in samples of a wild solanaceous plant Solanum umbelliferum from California, U.S., and called haplotype G (Mauck et al. 2019).
Lso is an unculturable bacterium, and thus the different strains of this species have only been defined by genetic markers. The different haplotypes, which can be seen as groups of closely related genotypes, are defined by the sequence of 16S rRNA and the sequence of 50S ribosomal protein rplJ/rplL gene region (Nelson et al. 2011, 2013). To distinguish the different strains within a haplotype, a multi-locus sequence typing (MLST) scheme based on seven protein-coding genes was developed (Haapalainen et al. 2018). These genetic markers can be used for the identification of the haplotype and strain of the Lso found in a plant or insect sample and also for characterizing previously unknown strains.
Since the first discovery of Lso in symptomatic carrots in Finland (Munyaneza et al. 2010), plant samples have been collected from carrot fields every year and the presence of Lso tested by PCR (Haapalainen et al. 2017, 2018). Recently, Lso haplotype C was also detected in parsnips grown beside carrots in Tavastia Proper region (Haapalainen et al. 2018). To assess the frequency of Lso infections in the parsnips grown close to the carrot fields where symptomatic plants had been observed, a survey on parsnip fields was carried out in the autumn 2018. The genotype of the bacterium was determined for selected Lso positive plant samples. However, for some of the samples the marker DNA sequences obtained did not match with any of the known Lso haplotypes. In this study, we characterized these novel Lso genotypes and determined their phylogenetic relationship to the genotypes previously described.
Materials and methods
Plant samples
In autumn 2018, samples of carrots (Daucus carota ssp. sativus) and parsnips (Pastinaca sativa) showing symptoms of foliar discolouration were collected for Lso testing from different regions of Finland. Carrot samples were collected from ten fields in South Savo, six fields in Satakunta and two fields in Southwest Finland. Parsnip samples were collected from three fields in South Savo and from five fields in Satakunta, at locations close to carrot fields. In addition, samples of weeds with foliar discolouration were collected from one carrot field and one parsnip field in Satakunta. Single Lso positive carrot samples from South Savo and weed samples from Tavastia Proper that had been collected in the earlier surveys in 2013 and 2016 and given unfamiliar rplJ/rplL PCR products were included in the sequence analyses.
DNA extraction, amplification and sequencing
For DNA extraction, 100 mg samples were cut from the petioles of the plants collected and these samples were homogenized in FastPrep tubes with lysing matrix A (MP Biomedicals, Santa Ana, CA, USA), using the FastPrep device at speed 5 for 40 s. DNA extraction was subsequently performed using the DNeasy Plant Mini kit protocol (Qiagen, Hilden, Germany), with DNA elution in 100 μl of nuclease-free water. For Lso detection, either end-point PCR with 16S primers OA2/Lsc2 (Haapalainen et al. 2017; Liefting et al. 2009) or real-time PCR with fluorescent probe (Li et al. 2006, 2009) was used as previously described (Haapalainen et al. 2017, 2018). For analysing the Lso genotype in the positive samples, end-point PCR was run with the ribosomal rplJ/rplL gene region primers rp01F(CL514-F)/rp01R(CL514-R) (Liefting et al. 2009) and with the primers amplifying fragments of the seven protein-coding genes - adk, atpA, fbpA, ftsZ, glyA, groEL and gyrB - included in the multi-locus sequence typing (MLST) scheme (Haapalainen et al. 2018). For a sample found to contain a novel haplotype of Lso also the 16S–23S intergenic spacer region was amplified with primers LpFrag4-1611F and LpFrag4-480R (Hansen et al. 2008). The PCR products were purified using QIAquick PCR purification kit (Qiagen) according to the manufacturer’s instructions. Capillary sequencing was performed at the Jokioinen laboratory of the Natural Resources Institute Finland and at the FIMM sequencing service of the University of Helsinki, both using an automated sequencer (Applied Biosystems, Waltham, MA, USA).
Phylogenetic analysis
After removing the primer sequences and trimming the 5′ ends, the sequences of the rplJ/rplL gene region were aligned by Clustal W and the alignment was then corrected based on the translated amino acid sequences of RplJ and RplL. A maximum parsimony (MP) phylogenetic tree was constructed in MEGA7 (Kumar et al. 2016), treating the gaps as the fifth character state, and bootstrapping with 1000 replications. The 16S rRNA gene sequences and the 16S–23S intergenic spacer regions were also aligned by Clustal W in MEGA7 and the small nuclear polymorphisms searched. The seven MLST gene fragments were aligned after primer trimming and then concatenated, forming a supermatrix, and used for pair-wise comparisons of the different sequence types as previously described (Haapalainen et al. 2018). To include the shorter haplotype G sequences in the analysis, all the gene fragments were trimmed at the 5′ end accordingly. Due to the lack of published sequence material, the haplotypes E and F could not be included in the MLST analyses. Phylogenetic distance matrixes were calculated using the substitution models TN93 (Tamura and Nei 1993), F84 (Felsenstein 1984) and K80 (Kimura 1980), and clustering was performed by unweighted-pair group method with arithmetic means (UPGMA). The maximum-likelihood tree was constructed using RAxML (Randomized Axelerated Maximum Likelihood) program v8.2.11 (Stamatakis et al. 2005) and applying GTR + G model to each partition (locus alignment). A majority rule (50%) consensus tree was computed out of 1000 bootstrap replicates.
Results and discussion
A novel haplotype of Lso in carrots and parsnips in eastern Finland
A previously unknown haplotype of the plant pathogen ‘Candidatus Liberibacter solanacearum’ was found in symptomatic cultivated carrots and parsnips in South Savo, eastern Finland. Two carrot plants infected with this haplotype, from now on called haplotype H, were collected as samples in the same locality within South Savo in 2013 and 2016 (Table 1; Fig. 1). In 2018, carrots infected with Lso at low bacterial titres (Ct values from 36 to 38) were found in seven of the ten fields sampled in South Savo and for two of the fields the Lso haplotype could be confirmed and was found to be C. In all the carrot fields sampled in Satakunta and Southwest Finland regions the symptomatic carrots were infected with Lso haplotype C. In the previous study in 2013 and 2014 carrot samples had been collected for Lso tests in Tavastia Proper, Ostrobothnia and South Savo regions in addition to Satakunta and Southwest Finland, and plants infected with Lso haplotype C had been found (Haapalainen et al. 2017). Thus, haplotype C is the major haplotype of Lso occurring in carrots in Finland and haplotype H is rare. In 2018, a survey on the occurrence of Lso in parsnips was carried out. Parsnips infected with Lso were found in all the three fields sampled in South Savo, and for two of these fields the Lso haplotype was identified as H. In the first field, four out of five samples were Lso positive and in the second field, 7.2 km distant from the first one, all five were Lso positive. In the third field three out of six samples were Lso positive, but due to the very low bacterial titre (real-time PCR Ct value >37) the haplotype could not be resolved. In contrast, the symptomatic parsnips collected in Tavastia Proper in 2016 (Haapalainen et al. 2018) and in Satakunta region in 2018 were infected with haplotype C. Thus, in these apiaceous crop plants the haplotype H has thus far only been detected within one sampling area located in eastern Finland.
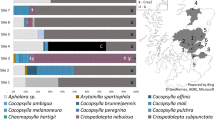
Regions in Finland where plants infected with ‘Candidatus Liberibacter solanacearum’ (Lso) were found. Lso haplotype C (diamonds) was detected in carrot and parsnip samples from Satakunta, Tavastia Proper, southwest Finland, south Savo and in the previous study (Haapalainen et al. 2017) also in carrots from South Ostrobothnia. Haplotype H was detected in carrots and parsnips in south Savo (solid circle) and in samples of polygonaceous plants from Satakunta and Tavastia Proper (open circles)
The carrots infected with Lso haplotype H showed strong foliar discolouration, similar to the symptom previously associated with a high titre of Lso haplotype C (Haapalainen et al. 2017; Nissinen et al. 2014). No proliferation of the shoot was observed, and the PCR test for phytoplasma with primers P1/fU5 and P7/rU3 (Crosslin et al. 2006) was negative. However, it is possible that other biotic or abiotic factors could have contributed to the discolouration symptom. In those Lso-infected parsnips that showed some symptoms, the symptoms were mild and included red discolouration of the petioles and slightly wrinkled leaves with blotchy bronze discolouration. Two of the plants (18-11P2 and 18-11P3) had brown discolouration in the root tip. No significant correlation was observed between the root/shoot weight ratio and the Lso titre in parsnips in this study: Spearman’s rank correlation coefficient was −0.193 with a p value 0.199. Previously, carrots infected with Lso haplotype C were shown to have a reduced root weight in a greenhouse experiment (Nissinen et al. 2014), whereas for the samples collected from fields the effect was not statistically significant, due to the differences between carrot varieties and growth conditions (Haapalainen et al. 2017). Furthermore, no shoot proliferation was observed in parsnips infected with Lso haplotypes C or H, in contrast to the severe symptoms associated with Lso haplotypes D and E (Alfaro-Fernández et al. 2017). Mild hairy root symptoms were occasionally detected in both Lso positive and Lso negative parsnips, suggesting other factors affecting the development of this symptom. The possible effects of Lso infection on the taste of the parsnip root have not been assessed.
Lso in Polygonaceae family weeds
In the family Polygonaceae, the species Fallopia convolvulus (wild buckwheat) and Persicaria lapathifolia (pale persicaria) are persistent and common annual weeds found in the fields and gardens and growing in the wild in all the regions of Finland. In 2016, one F. convolvulus and one P. lapathifolia growing as weeds within a carrot field in Tavastia Proper region tested positive for Lso. In 2018, three P. lapathifolia plants growing as weeds within a parsnip field and one F. convolvulus plant from a carrot field in Satakunta tested positive for Lso. The infected plants showed red discolouration (Fig. 2). The Lso positive F. convolvulus and P. lapathifolia plants found in 2016 and two of the Lso positive P. lapathifolia plants found in 2018 were infected with haplotype H (Table 1), whereas the third positive P. lapathifolia plant from the same parsnip field was infected with Lso haplotype C at a low titre (Ct value 35.76). The parsnips growing in that field were also infected with Lso haplotype C, sequence type 1. The one Lso positive F. convolvulus plant found from a carrot field in 2018 was infected with Lso haplotype C, similar to the symptomatic carrots taken as samples from the same location.
Transmission of Lso haplotypes C and H
Thus far, Lso haplotype H has not been detected in seed samples of carrot or other apiaceous plants (Haapalainen et al. 2018; Monger and Jeffries 2018), and thus it is unknown whether the infections with this haplotype in carrots and parsnips originate from the seed. In Finland the apiaceous crops are only grown as annual vegetables from imported seed. If Lso haplotype H does not occur or is very rare in the main areas of commercial carrot and parsnip seed production, then the infection source is more likely to be insect vectors.
Despite careful inspection, no adult psyllids, nymphs or eggs were detected on the plants infected with Lso haplotype H, and thus the vector(s) transmitting this haplotype is still unknown. However, since all the Ca. Liberibacter species and their haplotypes characterized thus far have been shown to be psyllid-transmitted, it is likely that the vector of haplotype H also belongs to the family Psyllidae. All the Lso-positive T. apicalis and T. anthrisci samples for which the haplotype was determined, contained Lso haplotype C (Haapalainen et al. 2018), and thus there might be another vector transmitting haplotype H. The finding in 2018 that within the same field the parsnips were infected with Lso haplotype C and the P. lapathifolia plants with haplotype H suggests that there could have been two different vectors with different host range. The absence of adult psyllids, nymphs or eggs on the infected parsnips suggests that parsnip is not a host plant of the new vector(s). Parsnip could, however, serve as an alternative host for T. apicalis and Lso haplotype C, if cultivated in crop rotation between carrot crops. Since T. apicalis showed clear oviposition preference for cultivated and wild carrot and coriander, while few eggs were laid on the other apiaceous plants (Valterová et al. 1997), it is unlikely that the carrot psyllids would prefer parsnip when both carrots and parsnips are cultivated during the same season. Psyllid species that occur in Finland and that could feed on P. lapathifolia are Aphalara borealis, A. freiji (syn. A. polygoni, Burckhardt and Lauterer 1997) and A. maculipennis (Ossiannilsson 1992). However, there is very little knowledge about their host plant range and preferences. According to Ossiannilsson (1992) they are strictly oligophagous on a few species belonging to Polygonum family, in which P. lapathifolia also was previously classified.
Genetic markers of the haplotype H
The 50S rplJ/rplL sequence was identical in all the eight haplotype H samples which were sequenced from this region. Previously, it was suggested that in the 50S rplJ/rplL sequence an alanine codon (GCT) had been inserted in the haplotype C rplL gene (Haapalainen et al. 2018). However, as haplotype H also has an alanine codon at this position and the haplotypes F and G and Ca. L. asiaticus have a valine codon (GTT) (Table 2; Supplementary File S1), it now seems more likely that Lso haplotypes A, B, D, E and U have lost one codon from rplL. In comparison with haplotype C ST1, haplotype H has six nucleotide differences in the 16S rRNA 1168 nt internal fragment (Table 2; Supplementary File S2) and five nucleotide differences in the 16S–23S intergenic spacer region (Table 2; Supplementary File S3).
Analysis of the seven protein-coding sequences of the multi-locus sequence typing (MLST) scheme revealed one polymorphic site (252: T or G) in the ftsZ gene fragment and one polymorphic site (155: G or A) in the gyrB gene fragment between the haplotype H samples. The G > T change in ftsZ turns an aspartate codon GAT into a tyrosine codon TAT, whereas the nucleotide change at the site 155 in gyrB is silent. All the four different combinations of these two SNPs were found amongst the eight sequenced samples, which were accordingly divided into four different sequence types (STs) (Table 1). The other five MLST gene fragments were identical in these eight samples (Supplementary File S4). The finding of four STs (bacterial “strains”) from such a small number of samples suggests that either the different STs have fairly equal frequencies or that the sampling was exceptionally lucky. The different alleles of ftsZ and gyrB do not seem to be linked to either western or eastern geographical location or to a certain plant species. This kind of pattern suggests that these STs have been present in Finland for a long time. For the Lso positive samples 18-11P2, 18-11P3, 18-11P4 and 18-11P8 only the ftsZ marker was sequenced. The results indicated that they also contained Lso haplotype H and that in 18-11P2, 18-11P3 and 18-11P4 the bacteria probably represented either ST1 or ST2 and in 18-11P8 either ST3 or ST4 (Table 1). Interestingly, the samples 18-11P7 and 18-11P8 that had G at the site 252 in ftsZ were collected from the same field, whereas the samples from 18-11P1 to 18-11P4 that had the SNP G > T were collected from another field. Thus, it is possible that the different fields had been visited by vectors belonging to different populations, harbouring different strains of Lso haplotype H.
The groEL PCR product of haplotype H showed one base change at the reverse primer groEL-1R binding site, where the first nucleotide was C instead of the A found in the other Lso haplotypes. Surprisingly, despite this mismatch, the reverse primer groEL-1R was still able to produce the expected 634 bp PCR product together with the forward primer groEL-1F. This SNP, a silent mutation in the leucine codon 341, was confirmed by performing a new set of PCR and sequencing reactions with alternative primers groEL-F (5’-TAGCAACTATTTCTGCTAACGG-3′) and groEL-R (5’-CTCTTTAAGCTTATCACGGTCA-3′).
Phylogeny of the Lso haplotypes
Based on the 50S ribosomal protein gene region rplJ/rplL and on the concatenated sequences of the seven MLST gene fragments, haplotype H appears distinct from the previously characterized haplotypes of Lso, which cluster together in phylogenetic analyses (Figs. 3, 4 and 5). The haplotype B that was seen as an outlier in previous analyses (Haapalainen et al. 2018; Hajri et al. 2017, 2019) appears to cluster together with the recently characterized haplotypes F and G. Separate from them, the haplotypes A, U, D and E form a cluster of closely related haplotypes. The genetic diversity within the haplotypes D and E was recently studied in detail (Hajri et al. 2019). As the haplotype A was originally found west of the mountain divide of North America (Nelson et al. 2011), its close relationship to three haplotypes found in Europe suggests that the ancestor of this cluster was distributed over an area including both north-western America and parts of Eurasia, connected by the Beringian land bridge. Haplotype C is closely related to this cluster. The new outlier is the haplotype H, which seems to represent an early divergent lineage of Lso. The three main branches of the Lso phylogeny are best visualized in the maximum likelihood tree based on the MLST sequences (Fig. 5). Thus, including the newly discovered haplotypes F (Swisher Grimm and Garczynski 2019), G (Mauck et al. 2019) and H in the phylogenetic analyses not only widens the family tree but also sharpens the picture of the relationships between the other haplotypes.
Maximum parsimony phylogenetic tree of the 50S rplJ/rplL gene region of ‘Candidatus Liberibacter solanacearum’ (Lso), with ‘Candidatus Liberibacter asiaticus’ (Las) as outgroup. The corresponding Lso haplotypes are indicated by letters on the right. The tree was obtained using the Subtree-Pruning-Regrafting (SPR) algorithm and the first tree out of the 10 most parsimonious trees (length = 177) is shown. The consistency index is 0.9306, the retention index 0.9606 and the composite index 0.8955. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test with 1000 replicates are shown next to the branches
Phylogenetic tree of the different multi-locus sequence types of ‘Candidatus Liberibacter solanacearum’ (Lso), based on unweighted-pair group method with arithmetic means (UPGMA) and on TN93 substitution model. The genomic sequences of ‘Ca. L. solanacearum’ used in the analysis were of haplotype A strains NZ1 and R1 (JMTK01000005.1 and GCA_000756225.1), haplotype B strain ZC1 (CP002371.1) and haplotype D strain ISR100 (GCA_002918245.2). Samples 13–12, 15-P24a, 15–167 and 15–214 represent haplotype C, sample 16–004 haplotype D and 15–108 haplotype U. Herbarium samples 51, 54, 59 and 61 represent haplotype G. Samples 13–356, 16-KT3, 18-11P7 and 18-T2 represent the novel haplotype H. The Lso haplotypes are indicated by letters on the right
Majority-rule consensus cladogram of ‘Candidatus Liberibacter solanacearum’ (Lso) and related (Candidatus) Liberibacter species constructed with RAxML (Randomized Axelerated Maximum Likelihood) program. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test with 1000 replicates are shown next to the branches. The genomic sequences used in the analysis were strains LcrBT0 and LcrBT1 (CP010522.1 and CP003789.1) for Liberibacter crescens, CLeuASNZ1 (PSQJ01000002.1) for ‘Ca. L. europaeus’, CLamSaoPaulo and CLamPW_SP (CP006604.1 and GCA_000350385.1) for ‘Ca. L. americanus’, CLafPTSAPSY (CP004021.1) for ‘Ca. L. africanus’, CLasIshi1 and CLasPsy62 (AP014595.1 and CP001677.5) for ‘Ca. L. asiaticus’, NZ1 and R1 (JMTK01000005.1 and GCA_000756225.1) for ‘Ca. L. solanacearum’ haplotype A, ZC1 (CP002371.1) for ‘Ca. L. solanacearum’ haplotype B and ISR100 (GCA_002918245.2) for ‘Ca. L. solanacearum’ haplotype D. Samples 13–12, 15-P24a, 15–167 and 15–214 represent haplotype C, sample 16–004 haplotype D and 15–108 haplotype U. Herbarium samples 51, 54, 59 and 61 represent haplotype G and samples 13–356, 16-KT3, 18-11P7 and 18-T2 represent the novel haplotype H. The Lso haplotypes are indicated by letters on the right
DNA sequences
The DNA sequences of Lso haplotype H 16S ribosomal RNA gene, 16S–23S intergenic region, 50S ribosomal protein gene region rplJ/rplL and the alleles of the seven MLST gene fragments of adk, atpA, fbpA, ftsZ, glyA, groEL and gyrB were deposited at GenBank with accession numbers from MK800158 to MK800169.
References
Alfaro-Fernández, A., Hernández-Llopis, D., & Font, M. I. (2017). Haplotypes of ‘Candidatus Liberibacter solanacearum’ identified in Umbeliferous crops in Spain. European Journal of Plant Pathology, 149, 127–131.
Antolinez, C. A., Fereres, A., & Moreno, A. (2017). Risk assessment of ‘Candidatus Liberibacter solanacearum’ transmission by the psyllids Bactericera trigonica and B. tremblayi from Apiaceae crops to potato. Scientific Reports, 7, 45534. https://doi.org/10.1038/srep45534.
Burckhardt, D., & Lauterer, P. (1997). Systematics and biology of the Aphalara exilis (Weber & Mohr) species assemblage (Hemiptera: Psyllidae). Entomologica Scandinavica, 28, 271–305.
Crosslin, J. M., Vandemark, G. J., & Munyaneza, J. E. (2006). Development of a real-time, quantitative PCR for detection of the Columbia Basin potato purple top phytoplasma in plants and beet leafhoppers. Plant Disease, 90, 663–667.
Felsenstein, J. (1984). Distance methods for inferring phylogenies: A justification. Evolution, 38, 16–24.
Haapalainen, M., Kivimäki, P., Latvala, S., Rastas, M., Hannukkala, A., Jauhiainen, L., et al. (2017). Frequency and occurrence of the carrot pathogen ‘Candidatus Liberibacter solanacearum’ haplotype C in Finland. Plant Patholology, 66, 559–570.
Haapalainen, M., Wang, J., Latvala, S., Lehtonen, M. T., Pirhonen, M., & Nissinen, A. I. (2018). Genetic variation of ‘Candidatus Liberibacter solanacearum’ haplotype C and characterization of a novel haplotype from Trioza urticae and stinging nettle. Phytopathology, 108(8), 925–934.
Hajri, A., Loiseau, M., Cousseau-Suhard, P., Renaudin, I., & Gentit, P. (2017). Genetic characterization of ‘Candidatus Liberibacter solanacearum’ haplotypes associated with apiaceous crops in France. Plant Disease, 101(8), 1383–1390.
Hajri, A., Cousseau-Suhard, P., Gentit, P., & Loiseau, M. (2019). New insights into the genetic diversity of the bacterial plant pathogen ‘Candidatus Liberibacter solanacearum’ as revealed by a new multilocus sequence analysis scheme. BioRxiv. https://doi.org/10.1101/623405.
Hansen, A. K., Trumble, J. T., Stouthamer, R., & Paine, T. D. (2008). A new Huanglongbing species, ‘Candidatus Liberibacter psyllauros’ found to infect tomato and potato, is vectored by the psyllid Bactericera cockerelli (Sulc). Applied and Environmental Microbiology, 74, 5862–5865.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 16(2), 111–120.
Kumar, S., Stecher, G., & Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33, 1870–1874.
Li, W., Hartung, J. S., & Levy, L. (2006). Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus huanglongbing. Journal of Microbiological Methods, 66, 104–115.
Li, W., Abad, J. A., French-Monar, R. D., Rascoe, J., Wen, A., Gudmestad, N. C., et al. (2009). Multiplex real-time PCR for detection, identification and quantification of ‘Candidatus Liberibacter solanacearum’ in potato plants with zebra chip. Journal of Microbiological Methods, 78, 59–65.
Liefting, L. W., Weir, B. S., Pennycook, S. R., & Clover, G. R. G. (2009). ‘Candidatus Liberibacter solanacearum’, a liberibacter associated with plants in the family Solanaceae. International Journal of Systematic and Evolutionary Microbiology, 59, 2274–2276.
Mauck, K. E., Sun, P., Meduri, V., & Hansen, A. K. (2019). New Ca. Liberibacter psyllaurous haplotype resurrected from a 49-year-old specimen of Solanum umbelliferum: A native host of the psyllid vector. Scientific Reports. https://doi.org/10.1038/s41598-019-45975-6.
Mawassi, M., Dror, O., Bar-Joseph, M., Piasezky, A., Sjölund, J. M., Levitzky, N., et al. (2018). ‘Candidatus Liberibacter solanacearum’ is tightly associated with carrot yellows symptoms in Israel and transmitted by the prevalent psyllid vector Bactericera trigonica. Phytopathology, 108, 1056–1066.
Monger, W. A., & Jeffries, C. J. (2018). A survey of ‘Candidatus Liberibacter solanacearum’ in historical seed from collections of carrot and related Apiaceae species. European Journal of Plant Pathology, 150, 803–815.
Munyaneza, J. E., Fisher, T. W., Sengoda, V. G., Garcynski, S. F., Nissinen, A., & Lemmetty, A. (2010). First report of “Candidatus Liberibacter solanacearum” associated with psyllid-affected carrots in Europe. Plant Disease, 94, 639.
Nelson, W. R., Fisher, T. J., & Munyaneza, J. E. (2011). Haplotypes of “Candidatus Liberibacter solanacearum” suggest long-standing separation. European Journal of Plant Pathology, 130, 5–12.
Nelson, W. R., Sengoda, V. G., Alfaro-Fernandez, A. O., Font, M. I., Crosslin, J. M., & Munyaneza, J. E. (2013). A new haplotype of “Candidatus Liberibacter solanacearum” identified in the Mediterranean region. European Journal of Plant Pathology, 135, 633–639.
Nissinen, A. I., Haapalainen, M., Jauhiainen, L., Lindman, M., & Pirhonen, M. (2014). Different symptoms in carrots caused by male and female carrot psyllid feeding and infection by ‘Candidatus Liberibacter solanacearum’. Plant Pathology, 63(4), 812–820.
Ossiannilsson, F. (1992). The Psylloidea (Homoptera) of Fennoscandia and Denmark. Fauna Entomologica Scandinavica, 26, 346 pp. Leiden (Netherlands): E. J. Brill.
Stamatakis, A., Ludwig, T., & Meier, H. (2005). RAxML-III: A fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics, 2, 456–463.
Swisher Grimm, K. D., & Garczynski, S. F. (2019). Identification of a new haplotype of ‘Candidatus Liberibacter solanacearum’ in Solanum tuberosum. Plant Disease, 103(3), 468–474.
Tahzima, R., Maes, M., Achbani, E. H., Swisher, K. D., Munyaneza, J. E., & De Jonghe, K. (2014). First report of ‘Candidatus Liberibacter solanacearum’ on carrot in Africa. Plant Disease, 98, 1426.
Tamura, K., & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10(3), 512–526.
Teresani, G. R., Bertolini, E., Alfaro-Fernandez, A., Martinez, C., Tanaka, F. A. O., Kitajima, E. W., et al. (2014). Association of ‘Candidatus Liberibacter solanacearum’ with a vegetative disorder of celery in Spain and development of a real-time PCR method for its detection. Phytopathology, 104, 804–811.
Valterová, I., Nehlin, G., & Borg-Karlson, A. K. (1997). Host plant chemistry and preferences in egg-laying Trioza apicalis (Homoptera, Psylloidea). Biochemical Systematics and Ecology, 25(6), 477–491.
Acknowledgements
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. The authors would like to thank Heikki Häkkinen (ELY Centre) for providing plant samples from South Savo, Marja Tuononen (Pro Agria) for her kind help in the sampling in 2018, Senja Räsänen, Ari Eskola and Outi Järvinen (Luke) for technical assistance and Anneli Virta (Luke) and Suvi Kyttänen (FIMM) for DNA sequencing.
Funding
Financial support for this study was provided by the Ministry of Agriculture and Forestry of Finland (project numbers 1842/03.01.02/2013 and 1506/03.01.02/2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Haapalainen, M., Latvala, S., Wickström, A. et al. A novel haplotype of ‘Candidatus Liberibacter solanacearum’ found in Apiaceae and Polygonaceae family plants. Eur J Plant Pathol 156, 413–423 (2020). https://doi.org/10.1007/s10658-019-01890-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01890-0