Abstract
Ramularia leaf spot (RLS) is a newly-important disease of barley which is caused when the fungus Ramularia collo-cygni enters necrotrophic development during colonisation of the host. Mutant alleles at the barley MILDEW LOCUS O, mlo, locus confer broad spectrum durable resistance against the powdery mildew fungus, Blumeria graminis f. sp. hordei, but can enhance susceptibility to pathogens with necrotrophic development stages such as R. collo-cygni. Given the importance of mlo in spring barley breeding programmes, identifying loci that mitigate the effect of mlo-mediated susceptibility on necrotrophic disease development is an important target. Mutation of the ENHANCED MAGNAPORTHE 1 (emr1) locus which can affect mlo-associated disease susceptibility, leads to a reduction in RLS symptoms on barley leaves but does not reduce R. collo-cygni accumulation. The effect of emr1 on the transition of R. collo-cygni from endophyte to necrotroph may relate to changes in reactive oxygen species in mutant plants which show reduced sensitivity to chloroplastic superoxide induced cell death and has lower relative chlorophyll content compared to mlo plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ramularia leaf spot (RLS) is an important disease of barley crops in many temperate countries (Havis et al. 2015; McGrann and Havis 2017). The disease is caused by the fungus Ramularia collo-cygni, which is transmitted through infected seed and by air-borne spores (Havis et al. 2014). R. collo-cygni has a long endophytic phase (Kaczmarek et al. 2017) with necrotrophic disease symptoms usually only visible once the crop has entered the reproductive phase (Schützendübel et al. 2008). Despite R. collo-cygni possessing the genetic capability to synthesise a range of secondary metabolites that could potentially affect disease severity (Dussart et al. 2018), expression of RLS appears to be linked with adverse environmental conditions (Makepeace et al. 2008; McGrann and Brown 2018; Peraldi et al. 2014). The vertical transmission by seeds and relationship between environmental conditions and symptom expression has led to the suggestion that R. collo-cygni may be an endophyte that only causes disease under specific circumstances (McGrann and Havis 2017). Resistance to RLS appears to be a quantitative trait although genetic studies in barley have indicated that mutant MILDEW LOCUS O (mlo) alleles increase susceptibility to RLS (McGrann et al. 2014), although this phenotype is not observed in all environments (Makepeace et al. 2007). Mutant mlo alleles are agriculturally important as they confer broad-spectrum resistance to all known field isolates of powdery mildew caused by Blumeria graminis f. sp. hordei in spring barley (Piffanelli et al. 2004). This resistance has been used to great effect for more than 40 years. The mutant allele is currently present in more than 70% of elite European varieties (Dreiseitl 2012; Jørgensen 1992) with most commercial spring barley with mlo-mediated mildew resistance carrying the mlo11 allele although some older varieties contain mlo9 (Jørgensen 1992).
The MLO gene encodes a seven-transmembrane domain protein (Büschges et al. 1997). Wild type MLO alleles are susceptibility factors for biotrophic powdery mildew fungi, which co-evolved before the divergence of monocot and dicot plants (Acevedo-Garcia et al. 2014; Appiano et al. 2015). However, introducing mlo in commercial spring barley varieties came with a number of undesirable pleiotropic effects. Plants containing mlo alleles developed spontaneous foliar necrosis spotting, tended to yield less (Kjaer et al. 1990; Wolter et al. 1993; Thomas et al. 1998) and were more susceptible to diseases with necrotrophic stages (Jarosch et al. 1999; Kumar et al. 2001; Jansen et al. 2005; McGrann et al. 2014). Plant breeding has managed to alleviate the necrotic spotting and yield penalties associated with mlo (Bjornstad and Aastveit 1990; Kjaer et al. 1990) but there are still concerns over increased disease susceptibility associated with this mutant allele. Mutant analyses have indicated that mlo-mediated susceptibility to the pathogens with necrotrophic growth stages is regulated by other genes. Two REQUIRED FOR MLO RESISTANCE (ROR1 and ROR2) genes are essential for the full expression of mlo-mediated resistance to powdery mildew (Freialdenhoven et al. 1996). Enhanced RLS symptom formation in mlo plants is reduced in ror1 and ror2 mutants although levels of R. collo-cygni DNA in colonised leaves are still elevated (McGrann et al. 2014). Mutant ror1 plants show decreased sensitivity to toxins produced by Bipolaris sorokiniana compared to mlo plants with the wild type ROR1 allele (Kumar et al. 2001). However, loss of ROR1 function has no effect on barley susceptibility to Magnaporthe oryzae (Jarosch et al. 1999) suggesting ROR genes may operate through mechanisms that differentially affect mlo-mediated susceptibility to specific fungal species. NEC1 encodes a cyclic nucleotide gated channel 4 protein, which when defective, results in a spontaneous lesion mimic phenotype in barley (Rostoks et al. 2006). Mutant nec1 alleles reduce lesion formation by Fusarium culmorum and R. collo-cygni in barley but these nec1-mediated phenotypes are differentially affected by mlo mutations. The presence of nec1 mutation in an mlo5 background was shown to have no effect on the development of RLS symptoms or the accumulation of R. collo-cygni DNA. However, the presence of mlo5 in a nec1 background compromises the reduction in F. culmorum lesion size usually seen in nec1 mutants, although disease levels did not reach those seen in wild type MLO or NEC1 plants or in mlo5 mutants (McGrann et al. 2015a). A screen for restoring resistance to M. oryzae in mlo-barley identified two loss of function mutations, ENHANCED MAGNAPORTHE RESISTANCE (EMR1 and EMR2), which showed partially restored resistance to M. oryzae with no compromise on mildew resistance (Jansen et al. 2007; Jansen and Schaffrath 2009). The gene responsible for the emr1 or emr2 phenotype have not yet been characterised. Genetic mapping of the emr1 locus indicated that the emr1 mutant contains at least one additional mutant gene, 3-KETOACYL-CoA SYNTHASE (HvKCS6), located on chromosome 4H that results in emr1 plants having reduced lower levels of cuticular waxes (Weidenbach et al. 2014). These mutagenesis studies have shown that the negative effect of mlo on certain pathogens with necrotrophic colonisation stages can be mitigated. Here, the effect of the emr1 mutation on RLS was examined to assess the potential of this locus to mitigate mlo-mediated susceptibility to this disease.
Materials and methods
Plant material
The emr1mlo5 mutant was derived from NaN3 mutagenesis of IngridBCmlo5 mutant (mlo5) and shown to restore resistance to the blast fungus, Magnaporthe oryzae, in the highly susceptible mlo5 background (Jansen et al. 2007). Experiments included the emr1mlo5 double mutant, the single mutant parental line mlo5 and the wild type donor cultivar of both mutants cv. Ingrid. Seeds were sown in to Levington M3 compost (ICL, Ipswich, UK) in rows of ten seeds per plastic tray (210 × 156 × 47 mm). Barley seedlings were cultivated in a MD1400 modular climate chamber (Snijders Labs, Tilberg, The Netherlands) at 18 °C with a 16:8 h light: dark cycle under 200 μmol m−2 s−1 artificial light at 80% relative humidity.
Ramularia collo-cygni isolates and inoculation method
The R. collo-cygni isolate DK05 Rcc001 (McGrann et al. 2016) collected from Denmark in 2005 was used in this study. Fungal cultures were stored on potato dextrose agar (PDA) plates supplemented with streptomycin (100 μg mL−1) at 15 °C. Liquid cultures were prepared by adding two 5 mm2 plugs from 14 to 28 day old PDA storage plates to 200 mL potato dextrose broth (PDB) supplemented with streptomycin (100 μg mL−1). PDB flasks with R. collo-cygni were incubated for 14 days at 15 °C in the dark on an orbital shaker set at 150 rpm. Barley seedlings were inoculated with R. collo-cygni as previously described (McGrann et al. 2014; Peraldi et al. 2014). Inoculum was prepared by fragmenting R. collo-cygni hyphal growth in the liquid cultures in a food processor (Kenwood Electronics, London, UK). A single drop of Tween20 (Sigma, Dorset, UK) was added to each batch of 50 mL inoculum that was used to inoculate approximately 240 barley plants at growth stage (GS) 12 (Zadoks et al. 1974). Plants were incubated in the dark for 48 h post inoculation and RLS lesions were scored as the proportion of the prophyll leaf covered with disease symptoms from 8 to 21 days post inoculation (dpi). Green leaf area (GLA) retention was assessed as the amount of prophyll leaf area not senescent at the same time point that RLS was scored in the emr1 mutant seedling inoculation experiments. A minimum of three independent inoculation experiments were assessed for each seedling inoculation experiment.
Field trials
Barley cv. Ingrid, the near isogenic line mlo5 and the emr1mlo5 mutant were sown in tussock plots at a trial site in Lanark, Scotland in 2015 and 2016. The Lanark site was selected as it typically exhibits high levels of R. collo-cygni (Havis et al. 2014). To maintain green leaf area retention and control other foliar diseases during the growing season the fungicides Comet® 200 (0.4 L ha−1 Pyraclostrobin [200 g L−1] BASF, Cheshire, UK) plus Vegas ® (0.25 L ha−1 Cyflufenamid [50 g L−1] Certis, Cambridgeshire, UK) were applied as a mixture to each plot at both growth stage (GS) 25–20 and GS45. RLS symptoms were scored 2–3 times after the crop had flowered (GS60 onwards) as the proportion of leaf F-1 covered with disease lesions.
Quantification of in planta Ramularia collo-cygni DNA levels
Genomic DNA (gDNA) was extracted from five leaves of cv. Ingrid, mlo5 and the emr1mlo5 mutant 21 dpi with R. collo-cygni isolate DK05 using the DNeasy Plant Mini Kit (Qiagen, Hildem, Germany) following the manufacturer’s protocol. R. collo-cygni DNA levels were quantified using a dilution series of R. collo-cygni DNA (50 ng to 0.1 pg) with qPCR following the method of Taylor et al. (2010). Data was collected from two independent inoculation experiments.
Dark-induced senescence assays
Differences in relative chlorophyll content and senescence between emr1mlo5, mlo5 and cv. Ingrid were tested using a SPAD 502 Plus Chlorophyll meter (Konica Minolta, Warrington, UK) in a dark-induced senescence assay as previously described (McGrann et al. 2015b). Relative chlorophyll content was taken from three points across the blade of GS12 prophyll leaves after excision from the plant (day 0). Leaves were placed on damp tissue paper in clear plastic boxes covered with aluminium foil and stored at room temperature. Additional measurements were taken every two days up to and including day eight of the experiment. Three independent experiments were examined with 6–8 leaves of each line tested in each experiment.
Sensitivity to reactive oxygen species-induced cell death
The sensitivity of emr1mlo5, mlo5 and cv. Ingrid to cell death induced by reactive oxygen species (ROS) was tested using detached prophyll leaves from barley plants at GS11–12 as described previously (McGrann et al. 2015b). Cell death was induced by the hydrogen peroxide donor alloxan (200 mM), the mitochondrial superoxide donor menadione (100 mM) and the chloroplastic superoxide donor methyl viologen (25 μM). Lesions were measured 96 h after treatment with each ROS donor using ImageJ software (Abràmoff et al. 2004). Three independent experiments were performed with eight leaves of each line assessed for each ROS donor in each replicate experiment.
Data analysis
RLS scores over each seeding inoculation time course experiment were used to calculate the area disease under progress curve (AUDPC; Shaner and Finney 1977). AUDPC data was converted into the percentage of the maximum AUDPC (%maxAUDPC) across the entire seedling inoculation time course experiment. Prior to analysis %maxAUDPC data was LOGIT+ transformed (McGrann et al. 2014) and then analysed using a general linear model (GLM). Differences in seedling levels of RLS %maxAUDPC and R. collo-cygni gDNA in seedling prophyll leaves 21 dpi of the emr1mlo5 mutant compared to mlo5 and Ingrid were tested using GLM with line and experiment as factors. Differences in GLA retention in emr1mlo5, mlo5 and cv. Ingrid was examined using linear mixed modelling of repeated measures. Differences in GLA retention were evaluated using the uniform correlation/split plot in time covariance matrix with dpi, line, experiment and the interactions between these factors set as fixed factors. The dpi by individual plant interaction term was set as the random factor in the model. Significant differences between mlo5 and emr1mlo5 or cv. Ingrid at specific dpi were assessed using a t-test. GLM was also used to assess the variation in RLS AUDPC in the field trials with line and year as factors following a Poisson transformation of the AUDPC data.
Variation in ROS-induced cell death between emr1mlo5, mlo5 and cv. Ingrid was assessed using a separate GLM for each ROS donor. Each GLM assessed the contribution of experiment, line and the interactions between these terms on the observed phenotypes. Differences in dark-induced senescence were tested using linear mixed modelling with repeated measurements as described above for GLA.
Results
Development of Ramularia leaf spot on the spring barley emr1mlo5 mutant
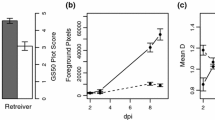
In seedling assays typical RLS symptoms were observed on leaves of barley cv. Ingrid, the mlo5 single mutant and the emr1mlo5 double mutant (Fig. 1a) 21 dpi with lesions first visible 8–10 dpi. The mlo5 mutant had increased RLS development compared to the wild type Ingrid (P < 0.001) as previously reported (McGrann et al. 2014, 2015a), whilst the emr1mlo5 mutant developed significantly less RLS than mlo5 (P < 0.001) but significantly more disease than cv. Ingrid (P = 0.007; Fig. 1b). By 8 dpi both mlo5 and emr1mlo5 had significantly less GLA than cv. Ingrid (P < 0.01) whereas emr1mlo5 plants had significantly lower GLA than mlo5 (P < 0.05) at this time point (Fig.1c). From 10 dpi onwards there were no significant differences in GLA between mlo5 and emr1mlo5 whereas both mutants had significantly lower levels of GLA compared to cv. Ingrid at all further time points (P < 0.001; Fig. 1c). Despite the differences in RLS development between emr1mlo5 and mlo5 there was no significant difference in the accumulation of R. collo-cygni gDNA 21 dpi between the mutants (P = 0.98) whereas cv. Ingrid leaves had significantly less fungal DNA than mlo5 (P = 0.022) and emr1mlo5 (P = 0.039) leaves (Fig. 1d).
Effect of emr1 mutation on Ramularia leaf spot (RLS) development. Disease phenotype on prophyll leaf of cv. Ingrid (MLO), mlo5 and emr1mlo5 plants 21 days post inoculation (dpi) with R. collo-cygni isolate DK05 Rcc001 (a). Development of RLS converted to the area under disease progress curve (AUDPC) and expressed as a proportion of the total AUDPC possible over the time course of the experiment (%maxAUDPC). Data was collected from five independent inoculation experiments with disease symptoms assessed on a minimum of eight plants of each line in each experiment (b). Green leaf area (GLA) retention over time following inoculation with R. collo-cygni isolate DK05 Rcc001. Data was collected from five independent inoculation experiments with GLA assessed on a minimum of eight plants of each line in each experiment (c). R. collo-cygni DNA levels in prophyll leaves 21 dpi measured by qPCR. Data was collected from two independent inoculation experiments with fungal DNA levels assessed from five replicates of each line in both experiments (d). Development of RLS in adult cv. Ingrid (MLO), mlo5 and emr1mlo5 plants. Field trials were conducted in two years with a single tussock plot scored for each line and the data from both years combined (e). Error bars indicate ±1 SE. ***P < 0.001; * 0.01 < P < 0.05 when emr1mlo5 or cv. Ingrid (MLO) plants are compared to mlo5
In field grown plants no significant differences in RLS development were recorded between the two years of the field trials (P = 0.37) nor were any differences observed between the lines (P = 0.74). However, even though the differences in RLS development between field grown cv. Ingrid, mlo5 and emr1mlo5 were not statistically significant there was a trend for mlo5 plants to exhibit more RLS symptoms than the other two lines (Fig. 1e).
Effect of emr1 mutation on relative chlorophyll content and dark-induced senescence
Processes linked with foliar senescence and chlorophyll breakdown have been linked with development of RLS (Schützendübel et al. 2008; McGrann et al. 2015b; McGrann and Brown 2018). Therefore, the effects of the emr1 mutation on senescence were examined using a dark-induced senescence assay. Prophyll leaves of the emr1mlo5 mutant at GS12 were a paler shade of green when compared to mlo5 or cv. Ingrid (Fig. 2a). This may be related to the altered wax status of emr1mlo5 plants (Weidenbach et al. 2014) or due to changes in chlorophyll content of the leaves associated with the emr1 mutation. Relative chlorophyll content of the emr1mlo5 mutant was significantly lower (P < 0.001) at day 0 than either mlo5 or cv. Ingrid. The relative chlorophyll content of emr1mlo5 remained significantly lower than mlo5 throughout the dark-induced senescence time course (day 2–6) but the SPAD readings for the two mutants were not significantly different by day 8 (Fig. 2b). Relative chlorophyll content of cv. Ingrid leaves was significantly higher than both emr1mlo5 (P < 0.001) and mlo5 (P < 0.05) across all time points.
Effect of the emr1 mutation on leaf phenotype and dark-induced senescence. Photograph of 14 day old prophyll leaf of cv. Ingrid (MLO), mlo5 and emr1mlo5 plants (a). Relative chlorophyll content for cv. Ingrid (MLO), mlo5 and emr1mlo5 plants during dark-induced senescence time course. Data was collected from three independent experiments with a minimum of six replicate plants tested in each experiment (b). Error bars indicate ±1 SE
Sensitivity of emr1mlo5 mutant to reactive oxygen species-induced cell death
Changes in ROS homeostasis associated with plant development and spontaneous cell death phenotypes have been linked with expression of RLS symptoms (McGrann and Brown 2018). No significant differences in the size of the alloxan-induced lesion (P = 0.49) were observed between emr1mlo5, mlo5 and cv. Ingrid (Fig. 3a+d). No significant difference in the size of the lesions formed by the mitochondrial superoxide donor menadione were recorded between the lines (Fig. 3b+e; P = 0.07) but the emr1mlo5 mutant did produce a significantly smaller lesion compared to mlo5 and cv. Ingrid (P < 0.001) when treated with the chloroplastic superoxide donor methyl viologen (Fig. 3c+f).
Sensitivity of emr1mlo5 mutant to reactive oxygen species (ROS)-induced cell death. Photographs of ROS-induced lesions on cv. Ingrid (MLO), mlo5 and emr1mlo5 prophyll leaves 96 h after treatment with alloxan (a), menadione (b) or methyl viologen (c). Measurements of lesion size following treatment with alloxan (d), menadione (e) or methyl viologen (f). Data was collected from three independent experiments with a minimum of five replicate plants tested in each experiment. Error bars indicate ±1 SE. ***P < 0.001 when emr1mlo5 or cv. Ingrid (MLO) plants are compared to mlo5
Discussion
Ramularia leaf spot has established itself as a serious threat to modern barley production across temperate climates (McGrann and Havis 2017; Havis et al. 2015; Walters et al. 2008). Yield losses typically range from 5 to 10% but can be as high as 70% without considering potential losses to grain quality which can reduce the value of the crop further (Havis et al. 2015). Resistance against RLS is most probably a quantitatively controlled genetic trait and the strong genotype by environment effects observed for the expression of disease symptoms between field trials, has led to problems in breeding effective resistance against this disease (Havis et al. 2015). Durable powdery mildew resistance conferred by the mlo locus has resulted in mildew not being the threat it once was (Makepeace et al. 2007) and the importance of this resistance source is clear by the presence of the mlo allele in approximately 70% of European spring barley varieties (Dreiseitl 2012; Jørgensen 1992). Even though mlo alleles can increase the threat of emerging diseases such as RLS (McGrann et al. 2014) the overriding importance of this mutation to spring barley production means that it is essential to develop varieties that are able to mitigate the negative effect of mlo on RLS (Brown and Rant 2013).
Limiting the effect of mlo on the necrotrophic phase of fungal diseases is an important breeding target to protect the use of mlo in commercial spring barley. Recent studies by Aghnoum et al. (2019) suggest that the genetic background of barley varieties can mitigate mlo-mediated susceptibility to blast disease. Through mutation analysis of an experimental mlo barley line, Jansen et al. (2007) demonstrated the presence of genetic loci that can moderate the effects of mlo on necrotrophic diseases. The mutant alleles, emr1 and emr2, both reduce mlo-mediated enhanced susceptibility to M. oryzae but have no effect on powdery mildew resistance (Jansen et al. 2007; Jansen and Schaffrath 2009). This suggests that there are genetic loci that have contrasting effects on the different pathways regulated by mlo, as previously suggested (McGrann et al. 2015a). Mutant emr1mlo5 plants showed reduced RLS development, but this reduction in disease was not accompanied by a reduction in fungal biomass. Similar findings were observed in barley ror mutants where both ror1 and ror2 reduce RLS expression in leaves but do not lower levels of R. collo-cygni biomass (McGrann et al. 2014). Not all genes that affect the expression of RLS interact with mlo. Mutant alleles of the cyclic nucleotide gated channel NEC1 lower RLS symptoms in wild type MLO plants but do not affect disease levels in a nec1 mlo double mutant (McGrann et al. 2015a) indicating that multiple pathways affect the expression of RLS symptoms and that mlo may only affect some of these pathways.
The emr1 mutant contains at least one other mutant locus, iwa1, which results in defective wax biosynthesis (Weidenbach et al. 2014). Whether or not altered surface wax composition affects RLS is currently unknown but emr1mlo5 mutants had reduced relative chlorophyll content and were more sensitive to chloroplastic ROS. Defects in ROR genes and NEC1 result in mis-regulated ROS and affect the transition of R. collo-cygni from asymptomatic growth to necrotrophic development. Changes in foliar chlorophyll content associated with the onset of senescence have been linked to RLS symptom expression (Schützendübel et al. 2008; McGrann and Brown 2018). Host responses associated with foliar senescence processes are regulated during R. collo-cygni infection suggesting that this pathogen triggers premature senescence in the plant (Sjökvist et al. 2019). Transgenic barley plants that have delayed leaf senescence due to over-expression of a Stress-induced NAC1 transcription factor show increased resistance against this disease (McGrann et al. 2015b). It is possible that defects in relative chlorophyll content, senescence and ROS balance in emr1mlo5 plants affect the transition of R. collo-cygni entering its necrotrophic phase. Determining the function of the gene at the EMR1 locus responsible for the effects on RLS symptom development may provide further insights into how plant genetics trigger this fungus to change growth habit and cause disease symptoms.
Whilst there is promise in modern commercial spring barley varieties for good RLS resistance in plants with mlo mildew resistance, a better understanding of the mechanisms and genetics controlling these phenotypes is required. Abiotic stress is known to affect expression of RLS (Peraldi et al. 2014; Makepeace et al. 2008; McGrann and Brown 2018) and Makepeace et al. (2007) suggested that the mlo effects on diseases such as RLS are influenced by environmental conditions. This finding is supported by the emr1mlo5 field trial data, where statistically significant differences between mlo, wild type MLO (cv. Ingrid) and emr1mlo5 were not recorded, despite the trend in RLS levels observed being similar to results obtained in the controlled environment tests. Assessing RLS development in varieties with good RLS resistance in mlo backgrounds in multiple environments is essential to confirm that the effect of genes moderating mlo-mediated susceptibility to RLS is not environmentally sensitive. This could be particularly important considering genetic loci such as EMR1, which are able to moderate the effect of mlo on RLS symptom expression, but do not appear to affect the accumulation of fungal biomass (McGrann et al. 2014, 2015a). As such R. collo-cygni DNA levels may remain high in spring barley varieties with mlo which have low levels of RLS. This could have negative consequences on grain quality and could result in epidemics when environmental conditions are favourable for the expression of RLS symptoms. The emr1 mutation has potential to mitigate the effect of mlo on promoting RLS symptom production in spring barley but further experimental evidence is needed to evaluate how the environment influences the effects of genetic loci that interaction with the mlo-response against this disease.
References
Abràmoff, M. D., Magalhães, P. J., & Ram, S. J. (2004). Image processing with imageJ. Biophotonics International, 11, 36–41.
Acevedo-Garcia, J., Kusch, S., & Panstruga, R. (2014). Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytologist, 204, 273–281.
Aghnoum, R., Bvindi, C., Menet, G., D’hoop, B., Maciel, J. L. N., & Niks, R. E. (2019). Host/nonhost status and genetics of resistance in barley against three pathotypes of Magnaporthe blast fungi. Euphytica, 215, 116.
Appiano, M., Catalano, D., Martínez, M. S., Lotti, C., Zheng, Z., Visser, R. G. F., Luigi, R., Yuling, B., & Stefano, P. (2015). Monocot and dicot MLO powdery mildew susceptibility factors are functionally conserved in spite of the evolution of class-specific molecular features. BMC Plant Biology, 15, 257.
Bjornstad, A., & Aastveit, K. (1990). Pleiotrophic effects on the mlo-o resistance gene in barley in different genetical backgrounds. Euphytica, 46, 217–226.
Brown, J. K. M., & Rant, J. C. (2013). Fitness costs and trade-offs of disease resistance and their consequences for breeding arable crops. Plant Pathology, 62(S1), 83–95.
Büschges, R., Hollricher, K., Panstruga, R., Simons, G., Wolter, M., Frijters, A., van Daelen, R., van der Lee, T., Diergaarde, P., Groenendijk, J., Töpsch, S., Vos, P., Salamini, F., & Schulze-Lefert, P. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell, 88, 695–705.
Dreiseitl, A. (2012). Frequency of powdery mildew resistances in spring barley cultivars in Czech variety trials. Plant Protection Science, 48, 17–20.
Dussart, F., Douglas, R., Sjökvist, E., Hoebe, P. N., Spoel, S. H., & McGrann, G. R. D. (2018). Genome-based discovery of polyketide-derived secondary metabolism pathways in the barley pathogen Ramularia collo-cygni. Molecular Plant-Microbe Interactions, 31, 962–975.
Freialdenhoven, A., Peterhansel, C., Kurth, J., Kreuzaler, F., & Schulze-Lefert, P. (1996). Ldentification of genes required for the function of non-race-specific. The Plant Cell, 8, 5–14.
Havis, N. D., Nyman, M., & Oxley, S. J. P. (2014). Evidence for seed transmission and symptomless growth of Ramularia collo-cygni in barley (Hordeum vulgare). Plant Pathology, 63, 929–936.
Havis, N. D., Brown, J. K. M., Clemente, G., Frei, P., Jedryczka, M., Kaczmarek, J., Kaczmarek, M., Matusinsky, P., McGrann, G. R. D., Pereyra, S., Piotrowska, M., Sghyer, H., Tellier, A., & Hess, M. (2015). Ramularia collo-cygni - an emerging pathogen of barley crops. Phytopathology, 105, 895–904.
Jansen, M., & Schaffrath, U. (2009). The barley mutant emr2 shows enhanced resistance against several fungal leaf pathogens. Plant Breeding, 128, 124–129.
Jansen, C., von Wettstein, D., Schäfer, W., Kogel, K.-H., Felk, A., & Maier, F. J. (2005). Infection patterns in barley and wheat spikes inoculated with wild-type and trichodiene synthase gene disrupted Fusarium graminearum. Proceedings of the National Academy of Sciences of the United States of America, 102, 16892–16897.
Jansen, M., Jarosch, B., & Schaffrath, U. (2007). The barley mutant emr1 exhibits restored resistance against Magnaporthe oryzae in the hypersusceptible mlo-genetic background. Planta, 225, 1381–1391.
Jarosch, B., Kogel, K., & Schaffrath, U. (1999). The ambivalence of the barley Mlo Locus : mutations conferring resistance against powdery mildew (Blumeria graminis f . sp . hordei ) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Molecular Plant-Microbe Interactions, 12, 508–514.
Jørgensen, I. H. (1992). Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica, 63, 141–152.
Kaczmarek, M., Piotrowska, M. J., Fountaine, J. M., Gorniak, K., McGrann, G. R. D., Armstrong, A., Wright, K. M., Newton, A. C., & Havis, N. D. (2017). Infection strategy of Ramularia collo-cygni and development of ramularia leaf spot on barley and alternative graminaceous hosts. Plant Pathology, 66, 45–55.
Kjaer, B., Jensen, H. P., Jensen, J., & Jørgensen, J. H. (1990). Associations between three ml-o powdery mildew resistance genes and agronomic traits in barley. Euphytica, 46, 185–193.
Kumar, J., Hückelhoven, R., Beckhove, U., Nagarajan, S., & Kogel, K. H. (2001). A compromised Mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (teleomorph: Cochliobolus sativus) and its toxins. Phytopathology, 91, 127–133.
Makepeace, J. C., Oxley, S. J. P., Havis, N. D., Hackett, R., Burke, J. I., & Brown, J. K. M. (2007). Associations between fungal and abiotic leaf spotting and the presence of mlo alleles in barley. Plant Pathology, 56, 934–942.
Makepeace, J. C., Havis, N. D., Burke, J. I., Oxley, S. J. P., & Brown, J. K. M. (2008). A method of inoculating barley seedlings with Ramularia collo-cygni. Plant Pathology, 57, 991–999.
McGrann, G. R. D., & Brown, J. K. M. (2018). The role of reactive oxygen in the development of Ramularia leaf spot disease in barley seedlings. Annals of Botany, 121, 415–430.
McGrann, G. R. D., & Havis, N. D. (2017). Ramularia leaf spot: A newly important threat to barley production. Outlooks on Pest Management, 28, 65–70.
McGrann, G. R. D., Stavrinides, A., Russell, J., Corbitt, M. M., Booth, A., Chartrain, L., Thomas, W. T. B., & Brown, J. K. M. (2014). A trade off between mlo resistance to powdery mildew and increased susceptibility of barley to a newly important disease, Ramularia leaf spot. Journal of Experimental Botany, 65, 1025–1037.
McGrann, G. R. D., Steed, A., Burt, C., Nicholson, P., & Brown, J. K. M. (2015a). Differential effects of lesion mimic mutants in barley on disease development by facultative pathogens. Journal of Experimental Botany, 66, 3417–3428.
McGrann, G. R. D., Steed, A., Burt, C., Goddard, R., Lachaux, C., Bansal, A., Corbitt, M., Gorniak, K., Nicholson, P., & Brown, J. K. M. (2015b). Contribution of the drought tolerance-related Stress-responsive NAC1 transcription factor to resistance of barley to Ramularia leaf spot. Molecular Plant Pathology, 16, 201–209.
McGrann, G. R. D., Andongabo, A., Sjökvist, E., Trivedi, U., Dussart, F., Kaczmarek, M., Mackenzie, A., Fountaine, J. M., Taylor, J. M. G., Paterson, L. J., Gorniak, K., Burnett, F., Kanyuka, K., Hammond-Kosack, K. E., Rudd, J. J., Blaxter, M., & Havis, N. D. (2016). The genome of the emerging barley pathogen Ramularia collo-cygni. BMC Genomics, 17, 584.
Peraldi, A., Griffe, L. L., Burt, C., McGrann, G. R. D., & Nicholson, P. (2014). Brachypodium distachyon exhibits compatible interactions with Oculimacula spp. and Ramularia collo-cygni, providing the first pathosystem model to study eyespot and ramularia leaf spot diseases. Plant Pathology, 63, 554–562.
Piffanelli, P., Ramsay, L., Waugh, R., Benabdelmouna, A., D’Hont, A., Hollricher, K., Jørgensen, J. H., Schulze-Lefert, P., & Panstruga, R. (2004). A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature, 430, 887–891.
Rostoks, N., Schmierer, D., Mudie, S., Drader, T., Brueggeman, R., Caldwell, D. G., Waugh, R., & Kleinhofs, A. (2006). Barley necrotic locus nec1 encodes the cyclic nucleotide-gated ion channel 4 homologous to the Arabidopsis HLM1. Molecular Genetics and Genomics, 275, 159–168.
Schützendübel, A., Stadler, M., Wallner, D., & Von Tiedemann, A. (2008). A hypothesis on physiological alterations during plant ontogenesis governing susceptibility of winter barley to ramularia leaf spot. Plant Pathology, 57, 518–526.
Shaner, G., & Finney, R. E. (1977). The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology, 77, 1051–1056.
Sjökvist, E., Lemcke, R., Kamble, M., Turner, F., Blaxter, M., Havis, N. D., Lyngkjær, M. F., & Radutoiu, S. (2019). Dissection of Ramularia leaf spot disease by integrated analysis of barley and Ramularia collo-cygni transcriptome responses. Molecular Plant-Microbe Interactions, 32, 176–193.
Taylor, J. M. G., Paterson, L. J., & Havis, N. D. (2010). A quantitative real-time PCR assay for the detection of Ramularia collo-cygni from barley (Hordeum vulgare). Letters in Applied Microbiology, 50, 493–499.
Thomas, W. T. B., Baird, E., Fuller, J. D., Lawrence, P., Young, G. R., Russell, J., Ramsay, L., Waugh, R., & Powell, W. (1998). Identification of a QTL decreasing yield in barley linked to Mlo powdery mildew resistance. Molecular Breeding, 4, 381–393.
Walters, D. R., Havis, N. D., & Oxley, S. J. P. (2008). Ramularia collo-cygni: The biology of an emerging pathogen of barley. FEMS Microbiology Letters, 279, 1–7.
Weidenbach, D., Jansen, M., Franke, R. B., Hensel, G., Weissgerber, W., Ulferts, S., Jansen, I., Schreiber, L., Korzun, V., Pontzen, R., Kumlehn, J., Pillen, K., & Schaffrath, U. (2014). Evolutionary conserved function of barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in providing wax signals for germination of powdery mildew fungi. Plant Physiology, 166, 1621–1633.
Wolter, M., Hollricher, K., Salamini, F., & Schulze-Lefert, P. (1993). The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Molecular & General Genetics, 239, 122–128.
Zadoks, J. C., Chang, T. T., & Konzak, C. F. (1974). A decimal code for the growth stages of cereals. Weed Research, 14, 415–421.
Acknowledgements
The authors thank Dr. Ulrich Schaffrath (RWTH Aachen, Germany) for providing the emr1 mutant and associated parental lines. SRUC receives funding from the Scottish Government RESAS.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by GMcG, SM and NDH. The first draft of the manuscript was written by GMcG and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Disclosure of potential conflicts of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
This article does not contain any research with human or animal subjects performed by any of the authors.
Informed consent
Not applicable.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
McGrann, G.R.D., Miller, S. & Havis, N.D. The ENHANCED MAGNAPORTHE RESISTANCE 1 locus affects Ramularia leaf spot development in barley. Eur J Plant Pathol 156, 123–132 (2020). https://doi.org/10.1007/s10658-019-01869-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01869-x