Abstract
Recent observations report the worldwide incidence of leaf-feeding grape phylloxera in formerly resistant scions of commercial vineyards. To analyze the genetic structure of leaf-feeding phylloxera, we performed an extensive sampling of leaf-feeding phylloxera populations in seven regions (“cantons”) in Switzerland and Germany. The use of polymorphic microsatellite markers revealed presence of 203 unique grape phylloxera multilocus genotypes. Genetic structure analyses showed a high genetic similitude of these European samples with phylloxera samples from its native habitat on Vitis riparia (northeastern America). Nevertheless, no genetic structure within the European samples was observed, and neither host, geography nor sampling date factors caused clear effects on phylloxera genetic stratification. Clonality was high in commercial vineyards and leaf-feeding grape phylloxera strains were found to be present in scion leaves and rootstock roots in the same vineyard, potentially indicating migration between both habitats. We found indications of sexual reproduction, as shown by high degrees of genetic variation among collection sites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Grape phylloxera (Daktulosphaira vitifoliae Fitch) is among the most important viticultural pests. The roots of the European grapevine (Vitis vinifera L.) are highly susceptible to this insect, which caused the devastation of many own-rooted vineyards when introduced into Europe in the nineteenth century. Farmers managed the pest by the subsequent use of partially-resistant American non-vinifera Vitis species or hybrids, as rootstocks. As an introduced pest, grape phylloxera can be commonly found nowadays in commercial vineyards, both feeding on roots of partially resistant rootstocks and leaves of partially resistant grapevine cultivars (Powell et al. 2013; Griesser et al. 2015). Recent reports show the incidence of leaf-feeding phylloxera on leaves of both partially resistant and formerly resistant vines (Bao et al. 2014; Fahrentrapp et al. 2015; Forneck et al. 2016). The vitality and lifespan of vineyards infested by root-feeding phylloxera (later on termed “phylloxerated”) depend on many factors, including rootstock origin, phylloxera biotype and population density, soil type, vineyard management practices and diverse abiotic and biotic stress factors (see Powell et al. 2013). Grape phylloxera can generate large economic losses to vine growers (Folwell et al. 2001), which emphasizes the need to identify and monitor strain diversity and seasonal population changes in commercial vineyards. Population genetic studies can provide insights into the evolution of reproduction modes, adaptive strategies of aphid species in agroecosystems, and the influence of environmental and anthropogenic factors on the genetic diversity and structure of aphid populations (Dixon 1977). Population genetic studies also facilitate the design and optimization of sustainable pest management strategies, such as effectively assessing and controlling grape phylloxera infestation levels (Benheim et al. 2012). They also play a key role in quarantine strategies (Clarke et al. 2017) and in the determination of vineyard value (Benheim et al. 2012).
Leaf-feeding phylloxera biotypes may cause damage in commercial vineyards, especially if phylloxera abundance is high early in the season. The phylloxera life cycle exists in many variants (reviewed in Forneck and Huber 2009). The most common life cycle in Europe begins in springtime with either the fundatrix hatching from an overwintering egg (holocycle) or by the first instar larva (hibernales) migrating from roots to leaves and inducing galls. During the grapevine vegetative cycle, both leaf- and root-feeding larvae reproduce asexually and reach up to 4–5 generations per season (Forneck et al. 2001). Toward the end of the season, alate nymphs produce sexual adults (sexuales) which mate, and a single egg (the overwintering egg) is laid by the female. Grape phylloxera feeds on partially-resistant rootstocks, establishing an infestation level that may affect the vigor and longevity of the vine, but rarely results in plant death (Benheim et al. 2012). Nevertheless, fatal plant effects have been occasionally observed in the case of infestation by phylloxera-devastating ‘superclones’, as found for ‘superclones’ G1 and G4 in Australian vineyards (Corrie et al. 2002). Phylloxerated vineyards may produce losses if only partially resistant rootstocks are chosen but, in general, root-feeding phylloxera is successfully managed by appropriate rootstock selection.
In regions where interspecific grape hybrids (V. vinifera x American Vitis species) are traditionally grown for either conventional or organic wine production (like Léon Millot or Maréchal Foch hybrids), high infestation rates on the leaves are frequently observed (Fahrentrapp et al. 2015; Jubb 1976). Prevailing reports also indicate a heavy incidence of leaf-galling phylloxera on V. vinifera cultivars, in an increasing random frequency throughout diverse winemaking regions worldwide. In this sense, phylloxeration has been observed in the canopies of some V. vinifera cultivars (e.g. Riesling, Chasselas, Chardonnay, Müller Thurgau, Cabernet Sauvignon and Viognier) in Germany (e.g. Forneck et al. 2017a), Switzerland (Fahrentrapp et al. 2015), Austria (Könnecke et al. 2010), Uruguay, Brazil, Peru (Vidart et al. 2013) and Australia (Powell, K.S. Pers. Comm.).
The reasons for the increasing infestation rates on leaves of V. vinifera cultivars and interspecific hybrids are unknown. Environmental factors, related to climate change conditions, as well as changes in vineyard management practices, have been discussed (Powell et al. 2003). The general decline of pesticide use in grape production, and changes towards intensive leaf management practices may provide a more favorable environment for leaf galling. Elevated soil temperatures and the lack of strong winter frost events may increase the survival of hibernating phylloxera instars leading to more abundant spring population densities. In years with early bud-break, the first reproducing generation of phylloxera have been observed by April in Austria (Forneck et al. 2017b pers. observation). As the number of generations increase, the population size expands both on leaves and roots, allowing establishment of multi-annual grape phylloxera populations in commercial vineyards. Phylloxera populations capable of feeding on rootstock roots and scion leaves can be classified in a series of defined biotypes (A-G), based on phylloxera - host plant interaction (Forneck et al. 2016). To date it is unclear whether these populations rise from hybridization, mutation, or if they are the result of new introductions. Although the actual effects of leaf infestation in commercial vineyards have not been systematically analyzed yet, it is likely to cause long-term economic losses to grape growers by reducing crop yield or through potential negative effects on wine quality.
This study aims to evaluate, for the first time, the genetic structure of leaf-feeding grape phylloxera populations in commercial vineyards of Central Europe by extensively sampling phylloxera populations in diverse regions (“cantons”) of Switzerland and Germany. Here, we tested if there was a link between phylloxera populations and plant host and/or edafo-climatic conditions in the vineyards. Results also provide novel information on phylloxera mode of reproduction. Finally, we add further evidence on the origin of the grape phylloxera population present in Central Europe by comparing our genotypes with previously reported strains from natural and introduced ranges.
Material and methods
European grape phylloxera sampling in commercial vineyards
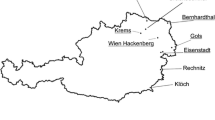
Leaf and soil emerged D. vitifoliae samples were collected as described previously (Fahrentrapp et al. 2015). In brief, commercial vineyards from 29 sampling sites throughout Switzerland and Germany wine-producing regions were selected (Fig. 1 and Table 1). Leaf gall samples were collected in 2013 and 2015 from May to October by detaching whole leaves from vineyard canopies and storing them at −20 °C until further analyses. Based on Powell et al. (2009), we used emergence traps for soil-emerging phylloxera collection, which were removed from soil after 2–4 weeks and rinsed with ethanol (70%). D. vitifoliae individuals were then collected and stored in ethanol at 2 °C until further processing. In total, 335 individuals were collected and considered in this study.
Sampling sites of grape phylloxera populations. 1: Nordweil; 2: Ebringen; 3: Batzenberg; 4: Hallau; 5: Rudolfingen; 6: Rafz; 7: Iselisberg; 8: Klingau; 9: Magden; 10: Maisprach; 11: Freienstein; 12: Dattlikon; 13: Aesch; 14: Reinach; 15: Leymen; 16: Regensberg; 17: Schinznach-Dorf; 18: Oberflachs; 19: Wildegg; 20: Quinten; 21: Twann; 22: Ligerz; 23: La Neuveville; 24: Sargans; 25: Malans; 26: Pully; 27: Nyon; 28: Malvaglia; 29: Novazzano
European grape phylloxera genotyping
Adult phylloxera individuals were individually finely ground with sterile plastic pestles in 200 μl of 5% Chelex BT 100 (BioRad, USA) solution for genomic DNA extraction. Ground samples were incubated at 90 °C for 20 min with frequent mixing, then thoroughly vortexed, centrifuged for 10 min, and 100 μl of the supernatant was transferred into a new tube. The Chelex BT-based extract was mixed with 10 μl of 3 M sodium acetate solution and 100 μl of isopropanol, and kept overnight at −20 °C for DNA precipitation. Samples were centrifuged at 500×g for 5 min, and DNA pellets were washed with 70% ethanol, followed by the addition of 100 μl of 1X TE buffer (Forneck et al. 2017a). A set of seven highly polymorphic SSR markers (Phy_III_55, Phy_III_30, Phy_III_36, Dvit6, DV4, DV8 and DVSSR4 (Forneck et al. 2017a)) were selected and analyzed in the 335 phylloxera samples. Polymerase chain reaction (PCR), separation of fragments and allele calling was performed following the procedures detailed in Riaz et al. (2017) and Forneck et al. (2017a). In every set of samples, six control genotypes were included to keep allele calling (Forneck et al. 2017a). The analysis revealed the presence of 203 unique grape phylloxera multilocus genotypes (MLGs).
Native grape phylloxera data
Available SSR data from 502 grape phylloxera MLGs were obtained from Lund et al. (2017) for comparison with international data. This dataset mainly corresponds to samples collected from grape phylloxera’s native range (USA), but also includes some samples from introduced ranges of Argentina (2), Brazil (3), California (5), Peru (3), Uruguay (4), Austria (7) and Hungary (8). Within the set of markers used to identify such MLGs, and to allow a joint analysis with the European samples, we focused on the analysis of 4 SSRs (Phy_III_55, Phy_III_30, Phy_III_36, Dvit6).
Data analysis
Phylloxera population structure was analyzed using the model-based clustering method implemented in STRUCTURE v.2.3 (Pritchard et al. 2000) in the whole set of unique European and American phylloxera MLGs. As stated above, this method was run on the basis of four SSR markers (Phy_III_55, Phy_III_30, Phy_III_36 and Dvit6) genotyped in both datasets, and assuming an admixture model with uncorrelated allele frequencies. The model was tested in a number of hypothetical genetic groups (K) ranging from 1 to 15, and each run was replicated 10 times to assess the consistency of the results, using a cycle of 250.000 burn-in steps followed by 500.000 Markov Chain Monte Carlo iterations. The most probable number of genetic groups was assessed following the ΔK criteria (Evanno et al. 2005) using STRUCTURE HARVESTER (Earl 2012). Phylloxera MLGs were assigned to a genetic group considering a membership coefficient over 0.95; otherwise, genotypes were considered as “admixed”. The same procedure was applied to the set of European MLGs, but using the seven SSR markers previously listed.
In parallel, a principal component analysis was performed by means of the DARwin software (Perrier and Jacquemoud-Collet 2006). Allele frequencies, mean number of alleles per locus, observed heterozygosity (Ho) and unbiased estimates of heterozygosity expected under Hardy-Weinberg assumptions (He) were calculated as previously indicated (Forneck et al. 2015). Clonal diversity (k) within populations was calculated for each population as k = G/N, where G is the number of different multilocus genotypes present in the sample and N is the sample size. Psex values were calculated with geneClone 2.0 software for every multicopy genotype in each population. Thresholds for Psex values were estimated for each population from Monte Carlo simulations. Significant Psex values indicate that multicopy genotypes are statistically overrepresented in a population and therefore, they are probably the result of clonal amplification. Fis values were included as a measure of inbreeding.
Results
Genetic structure: European vs. native grape phylloxera range
To generate a grouping according to descent, the 335 European samples obtained in this work were combined with a set of 502 phylloxera genotypes (470 native American genotypes and 32 phylloxera genotypes collected from various habitats in the introduced regions of Argentina (2), Austria (7), Brazil (3), California (5), Hungary (8), Peru (3), Uruguay (4)), previously analyzed in Lund et al. (2017). As a result, a global dataset of 837 phylloxera samples was created containing genetic information at four SSR loci. STRUCTURE analysis and ΔK criteria clearly suggested the most probable existence of two genetic groups (k1 and k2) within the 837 phylloxera samples analyzed. The majority (717 samples) were associated with one of two genetic groups (membership coefficient over 0.95), whereas 120 individuals were identified as “admixed”. According to this clustering, 99.7% of the individuals assigned to k1 were from North America (native habitat) from Vitis species like V. arizonica, V. vulpina, V. cinerea or V. labrusca, whereas the other genetic group (k2) was composed by the great majority of samples from the European commercial vineyards genotyped in this study (99.1%), samples from V. riparian host plants of the northeastern native range (sampled in Arizona, Indiana, Maine, New York, and Pennsylvania states), and most of the phylloxera genotypes sampled in diverse introduced regions (Fig. 2). This general grouping was confirmed by PCA results, where these two main genetic groups could be easily differentiated (Fig. 3).
Population structure of the 837 grape phylloxera samples included in this study based on STRUCTURE results. In A, every individual is shown as a vertical line, whose color indicates its origin: native range (yellow), Germany and Switzerland (brown) and introduced range (white) In B, individuals are also graphically represented by a vertical line, divided in colored segments according to the proportion of estimated membership in k1 (blue) and k2 (red). The optimal number of genetic groups (K = 2, k1 and k2) was established according to ΔK criteria. Accordingly, 305 and 412 samples are assigned to k1 and k2, respectively. Individuals from the native and introduced ranges were obtained from Lund et al. (2017)
Genetic diversity of European leaf-feeding grape phylloxera populations in commercial vineyards
After this general analysis, we focused on the detailed analysis of the 335 European grape phylloxera individuals obtained from commercial vineyards, which were genotyped by the use of seven SSR markers. Our aim was to analyze the genetic structure of these samples considering their geographical origin, host plant and feeding site (leaf/root), as shown in Table 1. In contrast to the strong stratification observed among native American vs. V. riparian + European samples, no population structure was observed when analyzing the European samples within the large range of the 15 hypothetical genetic groups tested, and no effect of geographical origin, feeding site (leaf/root) or host plant factors were identified. The latter result is consistent with those previously reported by Forneck et al. (2000) and Yvon and Peros (2003), who did not detect any significant effect of the host on phylloxera genotypes grouping.
Genetic data revealed high genetic diversity, yet most parameters indicate clonal propagation of the analyzed populations (Table 2). The average number of alleles per locus in the Swiss and German phylloxera populations ranged from 3.00 (Tessin) to 5.13 (Zürich). The observed heterozygosity of each subset of samples (Ho) ranged from 0.45 to 0.58, while the expected heterozygosity values (He) ranged from 0.50 to 0.58. For 9 of the 11 sites, He was greater than Ho. Tests of the Hardy-Weinberg equilibrium showed the presence of an excess of heterozygosity (Fis < 0) in some of the populations. The Fis values observed in the commercial vineyard populations are less (negatively) consistent within populations than those previously reported in studies performed in “semi-native habitats”, including abandoned rootstock areas with extensive leaf galling populations, research grapevine collections or rootstock nurseries (Forneck et al. 2015; Vorwerk and Forneck 2007). MLGs were observed in each sampling region (Supplement Table 1), indicating anholocyclic reproduction in commercial vineyards on both leaves and roots. The probability of independently produced repeated genotypes by sexual reproduction was determined through the calculation of Psex values in MLG simulations, obtaining values generally considered as low (Table 2). The individuals sampled in the Zürich region formed the largest population in our work (N = 110), and its analysis provided arguments (positive Fis values, highest allele frequencies, lowest Psex values) to support the existence of sexual reproduction events.
MLGs of leaf- vs. root-feeding populations
To elucidate the origin of the leaf-feeding grape phylloxera populations, we performed a detailed population genetic analysis on the MLGs identified therein. Sampling-site specific MLGs were detected in all the vineyards analyzed, and the list of the 28 MLGs with more than four repeats (out of five samples) are depicted in Supplement Table 1. Considering the whole set of individuals sampled (335), 40.3% (135 individuals) belonged to such MLGs.
To assess the existence of migrating MLGs among feeding habitats (leaves and roots) within vineyards, we focused on the analysis of samples from Zürich and Aargau, as these regions have been extensively studied (110 and 69 samples in Zürich and Aargau, respectively). MLGs 1, 2, 9 and 10 were found in both roots and leaves on grafted interspecific vines (Supplement Table 1). Other MLGs (15 and 16, or 4 and 5) were found to co-exist in the same vineyard, but they were found to feed separately on plant roots (MLGs 4 and 16) or leaves (MLGs 5 and 15). No migration was found between neighboring rootstock leaf habitats and either root- or leaf-feeding vineyards. For the first time, we report phylloxera MLGs migrating from root to leaf (or vice versa), with several examples in some Swiss commercial vineyards. For example, we found a commercial vineyard on Léon-Millot/unknown rootstock in Oberflachs (Aargau, sampling site AG40) with a high level of infestation (R = 18 vs. L = 5) in which a phylloxera MLG (MLG13) was present in both roots (4 samples) and leaves (5 samples). Similarly, in a Maréchal Foch/125AA (R = 5, L = 5) vineyard in Regensberg (ZH45, Zürich) we found that MLG26 was present in roots (4 samples) and leaves (4 samples). A third MLG (MLG44) was found both in the leaves (4 samples) and roots (1 sample) of a Léon Millot/unknown rootstock vineyard from Regensberg (ZH44).
Discussion
The likely patterns of the introductions of phylloxera into worldwide viticulture regions from its native habitat have been extensively analyzed by different authors using diverse genetic markers, including mitochondrial (Downie 2002) and nuclear polymorphisms (Forneck et al. 2000; Lund et al. 2017; Riaz et al. 2017). Whereas phylloxera populations from Vitis species like V. vulpina and V. arizonica are suggested to be the more likely source of introductions into viticulture regions like California (USA), South Africa, Australia, New Zealand and South America (Arancibia et al. 2018; Downie 2002; Lund et al. 2017), there is a general agreement that all European phylloxera introductions are likely to come from northeastern American populations, where V. riparia dominates (Downie 2002). Our results confirm such findings, in which most of the European genotypes from Swiss and German commercial vineyards are grouped together with phylloxera samples from the northeastern native range from V. riparia plants.
Aphid populations are composed of a small number of high frequency clones, and many low frequency (rare) genotypes (Harrison and Mondor 2011). In a recent report, Bao et al. (2014) have evaluated phylloxera genetic diversity in Uruguay through the screening of 75 leaf-feeding phylloxera from thirteen different regions (including semi-native habitats, nurseries and commercial vineyards), which were genotyped at four SSR loci. Similarly, Forneck et al. (2015) evaluated the genetic diversity of 315 leaf-feeding D. vitifoliae samples from semi-natural habitats throughout Austrian viticulture regions. Both studies show high degrees of genetic diversity, and population genetic parameters showed the predominant occurrence of asexual reproduction. In addition, none of these two works report the existence of phylloxera ‘superclones’ (MLGs with an outstanding capacity to predominate in a specific region and persist on time). This was expected, since the habitats chosen for these works were either semi-natural habitats (Forneck et al. 2015) or a mixture of nurseries, commercial vineyards or natural habitats (Bao et al. 2014), and thus not comparable to the suggested ‘superclone’ habitat (Vorburger et al. 2003). Here as well, no dominating MLG has been found (Supplement Table 1). As a result, no phylloxera leaf-feeding ‘superclone’ candidate has been identified within the Swiss and German commercial vineyards analyzed. Migration of phylloxera (based on genotypes) between locations has rarely been shown in semi-natural habitats in South America, Asia and Europe (e.g. Bao et al. 2014; Forneck et al. 2015; Sun et al. 2009; Vorwerk and Forneck 2007), and our results confirm these findings. No MLGs were sampled in multiple vineyards or regions, with the exception of MLG7, which was found in vineyards of Quinten (Graubünden) and Malvaglia (Tessin). Further studies aimed to analyze the local migration of phylloxera MLGs between adjacent plots by viticultural machinery and/or wind drift should be done to add evidence in short-distance dispersal mechanisms.
Previous population studies on D. vitifoliae in Europe showed two distinct genetic groups that correlated with their geographical location (northern and southern Europe), suggesting that selective forces could have favored the development of different phylloxera strains adapted to specific edafo-climatic conditions in northern and southern Europe (Forneck et al. 2000). The northern European group shows higher genetic diversity, and shows similitude with phylloxera genotypes from northeastern native habitats (Downie 2002). On the other hand, the southern European group has not been clustered to phylloxera genotypes sampled from a specific habitat. Here, all the sampling was performed above parallel 43°, in regions often referred to as the northern group (Forneck et al. 2000). As a result, we did not find any significant subgrouping among the European samples from commercial vineyard habitats, possibly due to the homogeneity of the environmental conditions of the regions sampled. Nevertheless, additional bottlenecks coming from planting strategies and distribution of (symptomless but infested) plant material as well as other further yet unknown factors may have affected phylloxera population structure.
Early introductions, together with rare sexual events, viticultural practices (plantings) or human-mediated transportation were responsible for the dissemination of phylloxera populations in the late nineteenth century in Europe, which lead to the adoption of new cultural practices in commercial vineyards (grafting). In recent years, potentially driven by diverse anthropogenic pressures (including global climate change effects and vineyard management systems), have caused an increment in the number of D. vitifoliae populations feeding on the leaves of grapevine scions. Here, we observed a high degree of genetic diversity within D. vitifoliae populations in commercial vineyards and we report that leaf-feeding D. vitifoliae in commercial vineyards can migrate from rootstock roots to scion leaves (and/or vice versa), establishing specific MLGs in each vineyard. Nonetheless, a dominating MLG (or ‘superclone’) has not been identified throughout the commercial vineyards from Switzerland and Germany analyzed in this work. No genetic structure was found within the European vineyards analyzed, and no conclusions regarding the impact of host plant (rootstock or scion), spatial range or elevation on phylloxera stratification could be made. This may be due to the limited set of samples, or the limited number of SSR markers used, which might have hampered the detection of effective alleles.
References
Arancibia, C., Riaz, S., Agüero, C., Ramirez-Corona, B., Alonso, R., Buscema, F., Martínez, L., & Walker, M. A. (2018). Grape phylloxera (Daktulosphaira vitifoliae Fitch) in Argentina: Ecological associations to diversity, population structure and reproductive mode. Australian Journal of Grape and Wine Research, 24, 284–291.
Bao, L. V., Scatoni, I. B., Gaggero, C., Gutiérrez, L., Monza, J., & Walker, M. A. (2014). Genetic diversity of grape phylloxera leaf galling populations on Vitis species in Uruguay. American Journal of Enology and Viticulture, 66(1), 46–53.
Benheim, D., Rochfort, S., Robertson, E., Potter, I., & Powell, K. S. (2012). Grape phylloxera (Daktulosphaira vitifoliae) – a review of potential detection and alternative management options. Annals of Applied Biology, 161(2), 91–115.
Clarke, C., Wigg, F., Norng, S., & Powell, K. S. (2017). Effectiveness of sodium hypochlorite as a disinfestation treatment against genetically diverse strains of grape phylloxera Daktulosphaira vitifoliae Fitch (Hemiptera: Phylloxeridae). Australian Journal of Grape and Wine Research, 23(3), 432–440.
Corrie, A. M., Crozier, R., Van Heeswijck, R., & Hoffmann, A. (2002). Clonal reproduction and population genetic structure of grape phylloxera, Daktulosphaira vitifoliae, in Australia. Heredity, 88(3), 203–211.
Dixon, A. (1977). Aphid ecology: life cycles, polymorphism, and population regulation. Annual Review of Ecology and Systematics, 8(1), 329–353.
Downie, D. (2002). Locating the sources of an invasive pest, grape phylloxera, using a mitochondrial DNA gene genealogy. Molecular Ecology, 11(10), 2013–2026.
Earl, D. A. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4(2), 359–361.
Evanno, G., Regnaut, S., & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology, 14(8), 2611–2620.
Fahrentrapp, J., Müller, L., & Schumacher, P. (2015). Is there need for leaf-galling grape phylloxera control? Presence and distribution of Daktulosphaira vitifoliae in Swiss vineyards. International Journal of Pest Management, 61(4), 340–345.
Folwell, R. J., Cifarelli, V., & Hinman, H. (2001). Economic consequences of phylloxera in cold climate wine grape production areas of eastern Washington. Small Fruits Review, 1(4), 3–15.
Forneck, A., & Huber, L. (2009). (A) sexual reproduction–a review of life cycles of grape phylloxera, Daktulosphaira vitifoliae. Entomologia Experimentalis et Applicata, 131(1), 1–10.
Forneck, A., Walker, M. A., & Blaich, R. (2000). Genetic structure of an introduced pest, grape phylloxera (Daktulosphaira vitifoliae Fitch), in Europe. Genome, 43(4), 669–678.
Forneck, A., Walker, M. A., & Blaich, R. (2001). An in vitro assessment of phylloxera (Daktulosphaira vitifoliae Fitch) (Hom., Phylloxeridae) life cycle. Journal of Applied Entomology, 125(8), 443–447.
Forneck, A., Anhalt, U. C., Mammerler, R., & Griesser, M. (2015). No evidence of superclones in leaf-feeding forms of Austrian grape phylloxera (Daktulosphaira vitifoliae). European Journal of Plant Pathology, 142(3), 441–448.
Forneck, A., Powell, K. S., & Walker, M. A. (2016). Scientific opinion: improving the definition of grape phylloxera biotypes and standardizing biotype screening protocols. American Journal of Enology and Viticulture, 67(4), 371–376.
Forneck, A., Dockner, V., Mammerler, R., Powell, K., Kocsis, L., Papura, D., et al. (2017a). PHYLLI–an international database for grape phylloxera (Daktulosphaira vitifoliae Fitch). IOBC-WPRS Bulletin, 128, 45–51.
Forneck, A., Eder, J., & Schmid, E. (2017b). Neue Gefahr: Blattgallen. Wissenschaftler warnen vor einer neuen Bedrohung durch die Reblaus in Zeiten des Klimawandels. Der Deutsche Weinbau, 23, 34–37.
Griesser, M., Lawo, N. C., Crespo-Martinez, S., Schoedl-Hummel, K., Wieczorek, K., Gorecka, M., Liebner, F., Zweckmair, T., Stralis Pavese, N., Kreil, D., & Forneck, A. (2015). Phylloxera (Daktulospharia vitifoliae) alters the carbohydrate metabolism in root galls to allowing the compatible interaction with grapevine (Vitis ssp.) roots. Plant Science, 234, 38–49.
Harrison, J. S., & Mondor, E. B. (2011). Evidence for an invasive aphid “superclone”: extremely low genetic diversity in oleander aphid (Aphis nerii) populations in the southern United States. PLoS One, 6(3), e17524.
Jubb, G., Jr. (1976). Grape phylloxera: incidence of foliage damage to wine grapes in Pennsylvania. Journal of Economic Entomology, 69(6), 763–766.
Könnecke, T., Aigner, C., Specht, S., Lawo, N., & Forneck, A. (2010). A stepwise assessment of Daktulosphaira vitifoliae infested grapevines in a Viennese vineyard site. Acta Horticulturae, 904, 59–62.
Lund, K. T., Riaz, S., & Walker, M. A. (2017). Population structure, diversity and reproductive mode of the grape phylloxera (Daktulosphaira vitifoliae) across its native range. PLoS One, 12(1), e0170678.
Perrier, X., & Jacquemoud-Collet, J. (2006). DARwin software: Dissimilarity analysis and representation for windows. Website http://darwin.cirad.fr/darwin. Accessed 1 Feb 2019.
Powell, K. S., Slattery, W. J., Deretic, J., Herbert, K., & Hetherington, S. (2003). Influence of soil type and climate on the population dynamics of grapevine phylloxera in Australia. Acta Horticulturae, 617, 33–41.
Powell, K. S., Korosi, G. A., & Mackie, A. M. (2009). Monitoring grape phylloxera populations using simple non-destructuve trapping systems. Acta Horticulture, 816, 29–34.
Powell, K. S., Cooper, P. D., & Forneck, A. (2013). The biology, physiology and host-plant interactions of grape phylloxera Daktulosphaira vitifoliae. Advances in Insect Physiology, 45, 159–218.
Pritchard, J., Stephens, M., & Donnley, P. (2000). Interference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Riaz, S., Lund, K. T., Granett, J., & Walker, M. A. (2017). Population diversity of grape phylloxera in California and evidence for sexual reproduction. American Journal of Enology and Viticulture, 68(2), 218–227.
Sun, Q.-H., Chen, Y.-C., Wang, H.-B., Downie, D., & Zhai, H. (2009). Origin and genetic diversity of grape phylloxera in China. Acta Entomologica Sinica, 52(8), 885–894.
Vidart, M. V., Mujica, M. V., Bao, L., Duarte, F., Bentancourt, C. M., Franco, J., & Scatoni, I. B. (2013). Life history and assessment of grapevine phylloxera leaf galling incidence on Vitis species in Uruguay. SpringerPlus, 2(1), 181.
Vorburger, C., Lancaster, M., & Sunnucks, P. (2003). Environmentally related patterns of reproductive modes in the aphid Myzus persicae and the predominance of two ‘superclones’ in Victoria, Australia. Molecular Ecology, 12(12), 3493–3504.
Vorwerk, S., & Forneck, A. (2007). Analysis of genetic variation within clonal lineages of grape phylloxera (Daktulosphaira vitifoliae Fitch) using AFLP fingerprinting and DNA sequencing. Genome, 50(7), 660–667.
Yvon, M., & Peros, J. (2003). Variation in aggressiveness and genetic diversity of grape phylloxera in southern France. Journal International des Sciences de la Vigne et du Vin, 37, 77–84.
Acknowledgements
Open access funding provided by University of Natural Resources and Life Sciences Vienna (BOKU). This study was financially supported by the FDW (Forschungsring des Deutschen Weinbaus) provided to MB. Further financial support was provided by Institute of Natural Resource Sciences of Zurich University of Applied Sciences. AF wishes to thank Markus Bauer for PHYLLI database set-up and support and Ulrike Anhalt, Jurrian Wilmink and Markus W. Eitle for discussion.
Funding
This study was funded by the FDW (Forschungsring des Deutschen Weinbaus), Germany and the Institute of Natural Resource Sciences of Zurich University of Applied Sciences, Switzerland.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Astrid Forneck declares that she has no conflict of interest.
Roswitha Mammerler declares that she has no conflict of interest.
Javier Tello declares that he has no conflict of interest.
Julia Müller declares that she has no conflict of interest.
Michaela Breuer declares that he has no conflict of interest.
Johannes Fahrentrapp declares that he has no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for.
the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Disclosure statement
No financial interests or benefits have arisen from the results reported in this work.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Forneck, A., Mammerler, R., Tello, J. et al. First European leaf-feeding grape phylloxera (Daktulosphaira vitifoliae Fitch) survey in Swiss and German commercial vineyards. Eur J Plant Pathol 154, 1029–1039 (2019). https://doi.org/10.1007/s10658-019-01723-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01723-0