Abstract
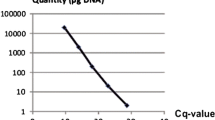
A real-time PCR assay was designed to quantify seed-borne infection of Pyrenophora graminea in barley (Hordeum vulgare). Conventional tests such as the freezing blotter method cannot distinguish P. graminea from the closely related P. teres. The seed infection threshold for P. graminea is lower than the one for P. teres and is therefore applied for both species although P. graminea may be absent. This results in unnecessary rejections of seed lots. PCR primers and a TaqMan probe were designed to target a P. graminea-specific DNA sequence. The potential of the real-time PCR assay for quantifying seed-borne infection of P. graminea was investigated by examining seed lots harvested from P. graminea-infected fields. The major part (84%) of the variation in the amount of P. graminea DNA measured by real-time PCR could be attributed to variation between seed lots while only about 8% was due to variation within seed lots. DNA quantities of P. graminea were positively correlated with seed infection incidence detected by the freezing blotter method as well as with the infection incidence of plants examined in the greenhouse. Both correlations were highly significant (P < 0.001) but the DNA quantities accounted only for 59% (R 2 = 0.59) and 56% (R 2 = 0.56), respectively, of the variation in the results obtained by the two conventional methods. Seed lots of varieties resistant to P. graminea contained considerable amounts of P. graminea DNA but showed no or only few leaf symptoms in the greenhouse test suggesting that the recommended seed infection thresholds could be raised for resistant varieties.



Similar content being viewed by others
References
Anonymous (2005). Sortsforsøg [Variety trials]. Landbrugets Rådgivningscenter. Landskontoret for Planteavl, Århus, Denmark.
Babadoost, M. (1997). Barley stripe. In D. E. Mathre (Ed.) Compendium of barley diseases ((pp. 24–25)2nd ed.). St. Paul, MN: The American Phytopathology Society.
Bates, J. A., & Taylor, E. J. A. (2001). Scorpion ARMS primers for SNP real-time PCR detection and quantification of Pyrenophora teres. Molecular Plant Pathology, 2, 275–280.
Bates, J. A., Taylor, E. J. A., Kenyon, D. M., & Thomas, J. E. (2001). The application of real-time PCR to the identification, detection and quantification of Pyrenophora species in barley seed. Molecular Plant Pathology, 2, 49–57.
Chilvers, M. I., du Toit, L. J., Akamatsu, H., & Peever, T. L. (2007). A real-time, quantitative PCR seed assay for Botrytis spp. that cause neck rot of onion. Plant Disease, 91, 599–608.
Cockerell, V., Kenyon, D. M., Mulholland, V., Bates, J. A., McNeil, M., Law, J. R., et al. (2004). Development of rapid seed health tests of Microdochium nivale seedling blight. Technical Paper no. 1: 18–44 HGCA project report no. 340, Home Grown Cereals Authority, London, UK.
Ghignone, S., & Migheli, Q. (2005). The database of PCR primers for phytopathogenic fungi. European Journal of Plant Pathology, 113, 107–109.
Husted, K. (1993). Detektion af plantepatogene svampe, eksemplificeret ved Pyrenophora graminea og Pyrenophora teres (with English summary). Ph.D. thesis, The Royal Veterinary and Agricultural University, Copenhagen.
Jørgensen, J. (1980). Comparative testing of barley seed for inoculum of Pyrenophora graminea and Pyrenophora teres in greenhouse and field. Seed Science and Technology, 8, 377–381.
Jørgensen, J. (1982a). The freezing blotter method in testing barley seed for inoculum of Pyrenophora graminea and P. teres. Repeatability of test results. Seed Science and Technology, 10, 639–646.
Jørgensen, J. (1982b). The freezing blotter method in testing barley seed for inoculum of Pyrenophora graminea. Varietal resistance and predictive value of test results. Seed Science and Technology, 10, 647–650.
McNeil, M., Roberts, A. M. I., Cockerell, V., & Mulholland, V. (2004). Real-time PCR assay for quantification of Tilletia caries contamination of UK wheat seed. Plant Pathology, 53, 741–750.
Möller, E. M., Bahnweg, G., Sandermann, H., & Geiger, H. H. (1992). A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruitbodies and infected plant tissue. Nucleic Acids Research, 20, 6115–6116.
Nielsen, B. J. (2001). Tolerancer for forekomst af udsædsbårne sygdomme i økologisk såsæd. In B. J. Nielsen & L. Kristensen (Eds.), Forædling af korn og bælgsæd samt produktion af såsæd i økologisk jordbrug (pp. 51–63). FØJO rapport nr. 15, Denmark.
Nielsen, G. C. (2006). Kassationsprocenter som følge af udsædsbårne svampe i økologisk udsæd af korn og bælgsæd 1999–2005. Planteavlsorientering nr. 09–748. Landbrugets Rådgivningscenter, Århus, Denmark.
Petersen, G., & Seeberg, O. (1998). Molecular characterization and sequence polymorphism of the alcohol dehydrogenase 1 gene in Hordeum vulgare L. Euphytica, 102, 57–63.
Pinnschmidt, H. O., & Nielsen, B. J. (2006). Leaf stripe resistance of spring barley cultivars. DARCOFenews. Retrieved from http://www.darcof.dk/enews/newsmail/december_2006/leaf.html
Schaad, N. W., & Frederick, R. D. (2002). Real-time PCR and its application for rapid plant disease diagnostics. Canadian Journal of Plant Pathology, 24, 250–258.
Schena, L., Nigro, F., Ippolito, A., & Gallitelli, D. (2004). Real-time quantitative PCR: A new technology to detect and study phytopathogenic and antagonistic fungi. European Journal of Plant Pathology, 110, 893–908.
Skoropad, W. R., & Arny, D. C. (1956). Histologic expression of susceptibility and resistance in barley to strains of Helminthosporium gramineum. Phytopathology, 46, 289–292.
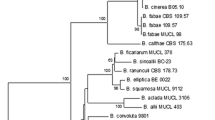
Smedegård-Petersen, V. (1983). Cross fertility and genetic relationship between Pyrenophora teres and P. graminea. The causes of net blotch and leaf stripe of barley. Seed Science and Technology, 11, 673–680.
Stevens, E. A., Blakemore, E. J. A., & Reeves, J. C. (1998). Relationships amongst barley and oat infecting isolates of Pyrenophora spp. Based on sequences of the internal. transcribed spacer regions of ribosomal DNA. Molecular Plant Pathology On-line. Retrieved from http://www.bspp.org.uk/mppol/1998/1111stevens
Taylor, E. J. A., Stevens, E. A., Bates, J. A., Morreale, G., Lee, D., Kenyon, D. M., et al. (2001). Rapid-cycle PCR detection of Pyrenophora graminea from barley seed. Plant Pathology, 50, 347–355.
Acknowledgement
This project was funded by the Danish Research Centre for Organic Farming (http://www.Darcof.dk). We would like to thank Bent Nielsen for supplying seed lots with P. graminea infection and those who supplied different Pyrenophora isolates for the primer specificity testing. We thank Dr. Kristian Kristensen for statistical advice and Henriette Nyskjold for expert technical assistance with the real-time PCR.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Justesen, A.F., Hansen, H.J. & Pinnschmidt, H.O. Quantification of Pyrenophora graminea in barley seed using real-time PCR. Eur J Plant Pathol 122, 253–263 (2008). https://doi.org/10.1007/s10658-008-9278-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-008-9278-1