Abstract
Adiposity, diabetes, and lifestyle factors are linked to gastroesophageal reflux disease (GERD) in observational studies. We conducted a two-sample Mendelian randomization analysis to determine whether those associations are causal. Independent genetic variants associated with body mass index (BMI), waist circumference (with and without adjustment for BMI), type 2 diabetes, smoking, and alcohol, coffee and caffeine consumption at the genome-wide significance level were selected as instrumental variables. Summary-level data for GERD were available from a genome-wide association meta-analysis of 71,522 GERD cases and 261,079 controls of European descent from the UK Biobank and QSkin Sun and Health studies. The odds ratio (OR) of GERD was 1.49 (95% confidence interval (CI), 1.40–1.60) for one standard deviation (SD) increase in BMI, 1.07 (95% CI, 1.04–1.10) for one-unit increase in log-transformed OR of type 2 diabetes, and 1.41 (95% CI, 1.31–1.52) for one SD increase in prevalence of smoking initiation. There were suggestive associations with GERD for higher genetically predicted waist circumference (OR per one SD increase, 1.14, 95% CI, 1.02–1.26) and caffeine consumption (OR per 80 mg increase, 1.08, 95% CI, 1.02–1.15). Genetically predicted waist circumference adjusted for BMI, alcohol or coffee consumption was not associated GERD. This study suggests causal roles of adiposity, diabetes, and smoking, and a possible role of high caffeine consumption in the development of GERD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastroesophageal reflux disease (GERD) is a common gastrointestinal disorder, affecting approximately 13% of the worldwide population and 20% of the adult population in the western countries [1, 2]. GERD impairs the patients’ life quality and increases the risk of other esophageal complications, such as esophagitis, esophageal strictures, Barrett esophagus, and esophageal adenocarcinoma [1, 2]. Epidemiological studies have revealed several possible risk factors for GERD, including excess adiposity [3,4,5,6,7], diabetes [8], smoking [6, 7, 9,10,11,12], alcohol consumption [11, 13], and coffee and caffeine consumption [6, 14]. However, evidence on most associations is equivocal with inconsistent findings across studies [15,16,17,18,19]. Additionally, unobserved confounding, misclassification, reverse causality, and other biases may hinder causal inference in these associations in observational studies. Determining the causal link of potentially modifiable risk factors with GERD is of great importance in understanding the etiology of this disease as well as in preventing and managing the disease in the clinical settings.
Mendelian randomization (MR) design uses genetic variants as instrumental variables for an exposure and can strengthen the causal inference [20]. This design can diminish residual confounding since the genetic variants are randomly assorted at conception and therefore have limited correlations with environmental and self-adopted factors [20]. In addition, the MR design can minimize the possibility of reverse causality because genetic variants cannot be modified by the development or progression of the disease [20]. A recent MR study in the UK Biobank study found an association for waist-to-hip ratio, but not for body mass index (BMI), smoking or caffeine consumption with risk of GERD, whereas the analyses for smoking and caffeinated-coffee consumption were underpowered [21]. Here, we conducted a two-sample MR study to examine the associations of overall and central adiposity, diabetes, smoking and alcohol, coffee, and caffeine consumption with risk of GERD based on more GERD cases and genetic instruments that explain more phenotypic variances.
Methods
Study design
Figure 1 shows the study design overview. This study was based on summary-level data on measures of adiposity, type 2 diabetes, lifestyle factors, and GERD from published genome-wide association studies (GWASs) and consortia. All studies included in the cited GWASs and consortia had been approved by a relevant review board and involved participants had given informed consent. The present MR analyses were approved by the Swedish Ethical Review Authority (2019‐02,793).
Study design overview. LD, linkage disequilibrium; SNPs, single-nucleotide polymorphisms. There are three assumptions of Mendelian randomization design. The first assumption is that the genetic variants used as instrumental variables should be robustly associated with the exposure; the second assumption is that the used genetic variants should not be associated with any confounders; and the third assumption is that the selected genetic variants should affect the risk of the outcome merely through the risk factor, not via alternative pathways
Instrument variable selection
Genetic variants (i.e., single nucleotide polymorphisms, SNPs) associated with BMI [22], waist circumference [23], type 2 diabetes [24], smoking initiation [25], and alcohol [25], coffee [26], and caffeine [27, 28] consumption at the genome-wide significance level (p < 5 × 10–8) were obtained from corresponding GWASs (Table 1). Smoking initiation was defined as a binary phenotype representing whether an individual had ever smoked cigarettes regularly (current or past smoker) [25]. SNPs associated with waist circumference adjusted for BMI was used to examine the BMI-independent effect of waist circumference [23]. Linkage disequilibrium among the SNPs was estimated using 1000 Genomes European panel as the reference population. Independent SNPs (i.e., SNPs without linkage disequilibrium, defined by r2 <0.001 and clumping window > 10,000 kb) were used as instrumental variables.
Gastroesophageal reflux disease data source
Summary-level data on the associations of exposure-related SNPs with GERD were obtained from a genome-wide association meta-analysis of the UK Biobank study and QSkin Sun and Health Study including a total of 71 522 GERD cases and 261 079 controls of European descent [29]. GERD cases were defined by filed codes of self-report, International Classification of Disease 9 and 10, the Office of Population Censuses and Surveys, and treatment/medicine in the UK Biobank study (68 535 cases and 250 910 controls), and self-reported heartburn and medical records of reflux medications in QSkin Sun and Health Study (2987 cases and 10 169 controls) [29]. The GWAS analysis was adjusted for recruitment age, genetic sex, the first ten principal components, and cryptic relatedness.
Statistical analysis
The inverse variance weighted method was used as the main statistical method (the random-effects model for the exposure constructed by ≥ 3 SNPs and the fixed-effect model for the exposure constructed by < 3 SNPs). Several sensitivity analyses, including the weighted median [30], MR-Egger [31], MR-PRESSO [32], and contamination mixture method [33] were conducted to examine the consistency of associations and detect and correct for horizontal pleiotropy. Assuming > 50% of weight from valid SNPs, the weighted median method can provide consistent estimates [30]. MR-Egger analysis can generate pleiotropy-corrected estimates if the intercept test detects significant horizontal pleiotropy (p for intercept < 0.05); however, the model is usually underpowered [31]. MR-PRESSO method can detect outlying SNPs and provide estimates after removal of outliers [32]. The embedded distortion test can examine the difference in estimates before and after removing outliers [32]. The contamination mixture method can provide robust estimates in analysis using hundreds of SNPs as instrumental variables with the presence of invalid SNPs [33]. To assess whether genetic liability to type 2 diabetes is associated with GERD risk independently of BMI, we performed the multivariable MR analysis with adjustment for genetically predicted BMI. Cochrane’s Q was used to assess the heterogeneity of estimates of SNPs. Power was estimated using an online tool (Table 1) [34]. Associations with p value < 0.007 (0.05/7 exposures) were deemed significant associations, and associations with a p value ≥ 0.007 and ≤ 0.05 were regarded as suggestive associations. All tests were two-sided and performed using the TwoSampleMR [35], MR-PRESSO [32] and MendelianRandomization [36] packages in the R software (version 4.0.2).
Results
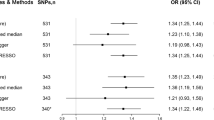
Higher genetically predicted BMI and genetic predisposition to type 2 diabetes (not diabetes diagnosis) and smoking initiation was associated with an increased risk of GERD (Fig. 2). The odds ratio (OR) of GERD was 1.49 (95% confidence interval (CI), 1.40, 1.60; p = 4.09 × 10–33) for one standard deviation (SD) increase in BMI, 1.07 (95% CI, 1.04, 1.10; p = 5.73 × 10–7) for one-unit increase in log-transformed OR of type 2 diabetes (not diabetes diagnosis), and 1.41 (95% CI, 1.31, 1.52; p = 2.12 × 10–19) for one SD increase in prevalence of smoking initiation. The association for type 2 diabetes slightly attenuated but remained significant in the multivariable MR analysis with adjustment for genetically predicted BMI (OR, 1.05, 95% CI, 1.03, 1.08; p = 2.76 × 10–4). There were suggestive associations for genetically predicted waist circumference (OR per one SD increase, 1.14, 95% CI, 1.02, 1.26; p = 0.017) and caffeine consumption (OR per 80 mg increase, 1.08, 95% CI, 1.02, 1.15; p = 0.013) (Fig. 2). The association for waist circumference attenuated in the analysis using SNP associated with waist circumference adjusted for BMI (OR per one SD increase, 1.08, 95% CI, 0.98, 1.18; p = 0.134) (Fig. 2). We did not observe any association of genetically predicted alcohol or coffee consumption with risk of GERD in the main analysis (Fig. 2).
Associations of genetically proxied adiposity, type 2 diabetes, and lifestyle factors with risk of gastroesophageal reflux disease. BMI, body mass index; CI, confidence interval; OR, odds ratio. Estimates were obtained from the inverse variance weighted method with random-effects with the except for estimate for caffeine consumption that was obtained from the inverse variance weighted method with fixed-effects.
Associations for all exposures remained overall consistent in sensitivity analyses (Table 2). The association became stronger for waist circumference adjusted for BMI in MR-Egger analysis and for waist circumference in the contamination mixture analysis (Table 2). Horizontal pleiotropy was observed in the MR-Egger analysis of BMI and type 2 diabetes (p for intercept < 0.05) (Table 2). One to ten outliers were detected by MR-PRESSO analyses; however, the associations remained consistent after removal of these outliers and no difference in estimates before and after removing outliers was observed (p for distortion test > 0.05) (Table 2).
Discussion
This MR study found that higher genetically predicted BMI and genetic liability to type 2 diabetes and smoking were associated with increased GERD risk. There were suggestive associations of genetically predicted higher waist circumference and caffeine consumption with an increased risk of GERD. The association for waist circumference attenuated after adjustment for genetically predicted BMI. Limited data was observed in support of an association of genetically predicted alcohol and coffee consumption with GERD.
Review articles of the association between obesity and GERD have found consistent evidence that overweight and obesity were associated with an ascended risk of GERD [3, 4, 17]. In a meta-analysis of 20 observational studies (mostly case–control and cross-sectional studies) with a total of 18 346 GERD patients, overweight and obesity was associated with a 57% and 115% higher risk of GERD, respectively [3], which is in line with our findings. This association is also supported by a cohort study including 29 610 Norwegians where weight loss was found to be associated with an increased odds of loss of GERD [5] and several randomized controlled trials [7]. However, a recent MR study of adiposity-related phenotypes in relation to GERD found that central adiposity, measured by waist-hip ratio instead of overall adiposity measured by BMI, showed a causal association with the increased risk of GERD [21]. Even though our study confirmed the positive association between central obesity measured by waist circumference and GERD, the association attenuated after adjustment for BMI. Thus, our data supported a stronger impact of overall adiposity, measured by BMI, compared to central adiposity on GERD. Several underlying mechanisms may explain the association between obesity and GERD, including esophageal motor abnormalities, low esophageal sphincter abnormalities, elevated intra-abdominal and intragastric pressures, increased frequency of transient sphincter relaxation, and esophageal inflammation [4, 37].
Gastrointestinal symptoms are frequently encountered in patients with type 2 diabetes. The association between type 2 diabetes and GERD was assessed in a meta-analysis including nine observational studies and the OR of GERD was 1.61 for diabetic individuals compared to non-diabetic controls [8]. Our MR study strengthened the causal nature of this positive association and further revealed that this association was independent of BMI. However, our MR estimate could not be compared with observational estimates since the risk of GERD was scaled to modelled liability to diabetes instead of diabetes diagnosis. Even though pathological pathways linking diabetes to GERD have not been fully investigated, the adverse effects of its upstream factors (e.g., obesity, smoking, etc.) on GERD and the autonomic neuropathy, especially vagal nerve damage in diabetic patients may explain this association [8, 38].
Evidence in support of an association between smoking and GERD was generally consistent [6, 7, 9,10,11,12]. A clinical trial in 14 smokers found that resuming smoking habits after abstaining from smoking for 48 h greatly increased acid reflux and heartburn [12]. The positive association between smoking and GERD was also revealed in case–control studies [11]. In The Trøndelag Health Study, smoking cessation was associated with an improvement in severe GERD in individuals with normal weight and anti-reflux medication, but not in individuals with minor GERD symptoms, overweight, or those using anti-reflux medication less than weekly [9]. Another study confirmed this association by comparing GERD improvement between the group that successful stopped smoking by varenicline and the group that failed to stop smoking [10]. Our MR study based on genetic data further strengthened the positive association between smoking and GERD even though we could not assess the interaction effects of body weight and the severity of the disease. A recent MR study did not observe this association, which might be caused by inadequate power [21]. Several mechanisms may decipher the increased risk of GERD in smokers, including reduced lower esophageal sphincter resting pressure (blocked cholinergic receptors by nicotine) and prolonged acid clearance time caused by reduced salivary secretion rate and bicarbonate concentration [39].
Consumptions of alcohol and coffee, two major beverages, have been associated with GERD risk with inconsistent or weak evidence for alcohol [11, 13, 15] and coffee consumption [6, 14, 15, 19]. A meta-analysis including 29 studies found an increased risk of GERD in regular alcohol drinkers compared to non- or occasional drinkers; however, this finding was majorly based on cross-sectional and case–control studies and therefore were prone to residual confounding from other alcohol intake-correlated lifestyles and behaviors as well as misclassification bias [13]. Our MR study did not observe a positive association between alcohol consumption and GERD risk, although we could not rule out the possibility that the observed null finding was led by an inadequate power or a possible association of heavy alcohol consumption or alcohol abuse with GERD. With regard to coffee consumption, our finding was in line with some [19] but not all [14] previous studies. Likewise, our null finding on coffee consumption is risked by an inadequate power. In a prospective analysis of data from Nurses’ Health Study II, consumption of coffee, tea, and soda (all contain caffeine) was associated with an increased risk of GERD symptoms, which may partly support the observed positive for caffeine consumption [6, 14], although the associations for coffee, tea, and soda in that study did not vary by caffeine status. Given that limited data examined the link from overall caffeine consumption to GERD risk, more study is needed.
Limitations need considerations when interpreting our results. The important limitation is the possible effect of horizontal pleiotropy. In this study, we observed significant pleiotropic effects in the analyses of BMI and type 2 diabetes, but not for other exposures. However, the associations for BMI and diabetes were consistent in sensitivity analyses. Anti-diabetes medications may be a pleiotropic source in the analysis of type 2 diabetes [40]. From the time logic (medications after disease onset), this is a vertical pleiotropy, which does not influence causal inference. Even though our analyses were based on a GERD dataset with a large sample size, we might overlook weak associations, especially for the exposure constructed by few SNPs that explain a small phenotypic variance. We had inadequate power in the analyses for waist circumference adjusted for BMI, and in the analyses of alcohol and coffee consumption. Our analysis included approximately 30% of GERD cases defined by self-reported information only in the UK Biobank, which might introduce outcome misclassification. However, the GERD genome-wide association study found strong genetic correlations across three GERD phenotypes defined by ICD10, self-reported GERD, and use of GERD medication, respectively (0.92 < rg < 0.97) [29], which indicated a good validity of self-report outcome data. Thus, the bias caused by self-report data should be minimal. Our study was confined to individuals of European descent, which reduced the population structure bias. On the other hand, this confinement may limit the generalizability of our findings to other populations. For smoking behaviors, the interaction effect with body weight and the severity of GERD could not be examined in this study based on summary-level data. For alcohol and coffee consumption, the effects of different types of alcohol or coffee could not be differentiated.
In summary, this MR study suggests causal roles of obesity, diabetes, and smoking in the development of GERD. The possible association between high caffeine consumption and an increased risk of GERD warrants confirmations.
Data availiability
The datasets analyzed in this study are publicly available summary statistics. Data used can be obtained upon a reasonable request to the corresponding author.
References
Richter JE, Rubenstein JH. Presentation and epidemiology of gastroesophageal reflux disease. Gastroenterology. 2018;154(2):267–76. https://doi.org/10.1053/j.gastro.2017.07.045.
Maret-Ouda J, Markar SR, Lagergren J. Gastroesophageal reflux disease: a review. JAMA. 2020;324(24):2536–47. https://doi.org/10.1001/jama.2020.21360.
Corley DA, Kubo A. Body mass index and gastroesophageal reflux disease: a systematic review and meta-analysis. Am J Gastroenterol. 2006;101(11):2619–28. https://doi.org/10.1111/j.1572-0241.2006.00849.x.
Hampel H, Abraham NS, El-Serag HB. Meta-analysis: obesity and the risk for gastroesophageal reflux disease and its complications. Ann Intern Med. 2005;143(3):199–211. https://doi.org/10.7326/0003-4819-143-3-200508020-00006.
Ness-Jensen E, Lindam A, Lagergren J, Hveem K (2013) Weight loss and reduction in gastroesophageal reflux: a prospective population-based cohort study the HUNT study. Am J Gastroenterol 108(3):376–82. https://doi.org/10.1038/ajg.2012.466
Mehta RS, Nguyen LH, Ma W, Staller K, Song M, Chan AT. Association of diet and lifestyle with the risk of gastroesophageal reflux disease symptoms in US women. JAMA Intern Med. 2021;181(4):552–4. https://doi.org/10.1001/jamainternmed.2020.7238.
Ness-Jensen E, Hveem K, El-Serag H, Lagergren J. Lifestyle intervention in gastroesophageal reflux disease. Clin Gastroenterol Hepatol. 2016;14(2):175–82. https://doi.org/10.1016/j.cgh.2015.04.176.
Sun XM, Tan JC, Zhu Y, Lin L. Association between diabetes mellitus and gastroesophageal reflux disease: A meta-analysis. World J Gastroenterol. 2015;21(10):3085–92. https://doi.org/10.3748/wjg.v21.i10.3085.
Ness-Jensen E, Lindam A, Lagergren J, Hveem K. Tobacco smoking cessation and improved gastroesophageal reflux: a prospective population-based cohort study: the HUNT study. Am J Gastroenterol. 2014;109(2):171–7. https://doi.org/10.1038/ajg.2013.414.
Kohata Y, Fujiwara Y, Watanabe T, et al. Long-term benefits of smoking cessation on gastroesophageal reflux disease and health-related quality of life. PLoS ONE. 2016;11(2): e0147860. https://doi.org/10.1371/journal.pone.0147860.
Watanabe Y, Fujiwara Y, Shiba M, et al. Cigarette smoking and alcohol consumption associated with gastro-oesophageal reflux disease in Japanese men. Scand J Gastroenterol. 2003;38(8):807–11. https://doi.org/10.1080/00365520310004506.
Kadakia SC, Kikendall JW, Maydonovitch C, Johnson LF. Effect of cigarette smoking on gastroesophageal reflux measured by 24-h ambulatory esophageal pH monitoring. Am J Gastroenterol. 1995;90(10):1785–90.
Pan J, Cen L, Chen W, Yu C, Li Y, Shen Z. Alcohol consumption and the risk of gastroesophageal reflux disease: a systematic review and meta-analysis. Alcohol Alcohol. 2019;54(1):62–9. https://doi.org/10.1093/alcalc/agy063.
Mehta RS, Song M, Staller K, Chan AT. Association between beverage intake and incidence of gastroesophageal reflux symptoms. Clin Gastroenterol Hepatol. 2020;18(10):2226-33.e4. https://doi.org/10.1016/j.cgh.2019.11.040.
Meining A, Classen M. The role of diet and lifestyle measures in the pathogenesis and treatment of gastroesophageal reflux disease. Am J Gastroenterol. 2000;95(10):2692–7. https://doi.org/10.1111/j.1572-0241.2000.03175.x.
Hunt R, Armstrong D, Katelaris P, et al. World gastroenterology organisation global guidelines: GERD global perspective on gastroesophageal reflux disease. J Clin Gastroenterol. 2017;51(6):467–78. https://doi.org/10.1097/mcg.0000000000000854.
Cheng Y, Kou F, Liu J, Dai Y, Li X, Li J. Systematic assessment of environmental factors for gastroesophageal reflux disease: an umbrella review of systematic reviews and meta-analyses. Dig Liver Dis. 2021;53(5):566–73. https://doi.org/10.1016/j.dld.2020.11.022.
Festi D, Scaioli E, Baldi F, et al. Body weight, lifestyle, dietary habits and gastroesophageal reflux disease. World J Gastroenterol. 2009;15(14):1690–701. https://doi.org/10.3748/wjg.15.1690.
Kim J, Oh SW, Myung SK, et al. Association between coffee intake and gastroesophageal reflux disease: a meta-analysis. Dis Esophagus. 2014;27(4):311–7. https://doi.org/10.1111/dote.12099.
Burgess S, Thompson SG. Mendelian randomization: methods for using genetic variants in causal estimation: CRC Press; 2015.
Green HD, Beaumont RN, Wood AR, et al. Genetic evidence that higher central adiposity causes gastro-oesophageal reflux disease: a Mendelian randomization study. Int J Epidemiol. 2020;49(4):1270–81. https://doi.org/10.1093/ije/dyaa082.
Pulit SL, Stoneman C, Morris AP, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166–74. https://doi.org/10.1093/hmg/ddy327.
Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–96. https://doi.org/10.1038/nature14132.
Vujkovic M, Keaton JM, Lynch JA, et al. (2020) Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet 52(7):680–91. https://doi.org/10.1038/s41588-020-0637-y
Liu M, Jiang Y, Wedow R, et al. (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet 51(2):237–44. https://doi.org/10.1038/s41588-018-0307-5
Zhong VW, Kuang A, Danning RD, et al. A genome-wide association study of bitter and sweet beverage consumption. Hum Mol Genet. 2019;28(14):2449–57. https://doi.org/10.1093/hmg/ddz061.
Cornelis MC, Kacprowski T, Menni C, et al. Genome-wide association study of caffeine metabolites provides new insights to caffeine metabolism and dietary caffeine-consumption behavior. Hum Mol Genet. 2016;25(24):5472–82. https://doi.org/10.1093/hmg/ddw334.
McMahon G, Taylor AE, Davey Smith G, Munafò MR. Phenotype refinement strengthens the association of AHR and CYP1A1 genotype with caffeine consumption. PLoS ONE. 2014;9(7): e103448. https://doi.org/10.1371/journal.pone.0103448.
An J, Gharahkhani P, Law MH, et al. Gastroesophageal reflux GWAS identifies risk loci that also associate with subsequent severe esophageal diseases. Nat Commun. 2019;10(1):4219. https://doi.org/10.1038/s41467-019-11968-2.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14. https://doi.org/10.1002/gepi.21965.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25. https://doi.org/10.1093/ije/dyv080.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. https://doi.org/10.1038/s41588-018-0099-7.
Burgess S, Foley CN, Allara E, Staley JR, Howson JMM. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun. 2020;11(1):376. https://doi.org/10.1038/s41467-019-14156-4.
Brion MJ, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501. https://doi.org/10.1093/ije/dyt179.
Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018. https://doi.org/10.7554/eLife.34408.
Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–9. https://doi.org/10.1093/ije/dyx034.
El-Serag HB, Thrift AP. Obesity and gastroesophageal reflux disease. The Esophagus 2021. pp. 624–32.
Verne GN, Sninsky CA. Diabetes and the gastrointestinal tract. Gastroenterol Clin North Am. 1998;27(4):861–74. https://doi.org/10.1016/s0889-8553(05)70035-2.
Ness-Jensen E, Lagergren J. Tobacco smoking, alcohol consumption and gastro-oesophageal reflux disease. Best Pract Res Clin Gastroenterol. 2017;31(5):501–8. https://doi.org/10.1016/j.bpg.2017.09.004.
Nishida T, Tsuji S, Tsujii M, et al. Gastroesophageal reflux disease related to diabetes: analysis of 241 cases with type 2 diabetes mellitus. J Gastroenterol Hepatol. 2004;19(3):258–65. https://doi.org/10.1111/j.1440-1746.2003.03288.x.
Acknowledgements
Genetic association estimates for gallstone disease were obtained from a genome-wide association meta-analysis of the UK Biobank study and QSkin health study cohort. The authors thank all investigators for sharing these data.
Funding
Open access funding provided by Karolinska Institute. The study is funded by the Swedish Research Council for Health, Working Life and Welfare (Forte; grant no. 2018–00123) and the Swedish Research Council (Vetenskapsrådet; grant no. 2019–00977). Funders had no roles in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
Author information
Authors and Affiliations
Contributions
Study conception and design: SY, SC.L; data acquisition and analysis: SY; drafting the manuscript and figures: SY; reviewing the manuscript: SY, SCL
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they donot have any conflict of interest to declare.
Ethical approval
All studies had been approved by a relevant ethical review board and participants had given informed consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yuan, S., Larsson, S.C. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a Mendelian randomization study. Eur J Epidemiol 37, 747–754 (2022). https://doi.org/10.1007/s10654-022-00842-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10654-022-00842-z