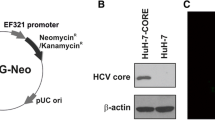
Abstract
Background
Hepatitis C virus infects ~3 % of the population and it is a risk factor for hepatocarcinogenesis. The epigenetic mechanisms of HCV-induced hepatocyte transformation towards malignancy in this context are unclear.
Aims
The purpose of this study was to evaluate the effect of HCV core proteins of different genotypes on DNA methyltransferases (DNMTs) induction.
Materials/Methods
We investigated DNMT1, DNMT3b and E-Cadherin (CDH1) mRNA and protein expression levels in an in vitro model of Huh-7 cells expressing the HCV core protein of different genotypes: 1b, 2a, 3a, 4h and 5a.
Results
We found that both mRNA and protein expression levels of DNMT1 and 3b were upregulated in genotype 1b HCV core expressing cells as compared to control cells. DNMT3b mRNA levels did not change in genotypes 2a, 3a, 4h and 5a, but were upregulated at the protein level by genotype 1b, 2a, 3a. CDH1 mRNA expression was downregulated only in genotype 1b, whereas its protein expression resulted in downregulation by the HCV core of genotypes 1b, 2a and 3a. Conversely, no significant changes were observed for DNMTs and CDH1 investigated in Huh-7 cells expressing the genotypes 4h and 5a. Furthermore, we present evidence that HCV core 1b protein expression induces DNMTs overexpression through STAT3 protein as demonstrated by NSC74859 treatment. Moreover, SIRT1 inhibition affected DNMT1 and 3b expression only in HCV core protein genotype 1b expressing cells as demonstrated by treatment with its inhibitor sirtinol.
Conclusions
Our findings suggest that HCV core protein could play a role in HCC development at least in part by altering DNMTs expression.




Similar content being viewed by others
References
Tellinghuisen TL, Rice CM. Interaction between hepatitis C virus proteins and host cell factors. Curr Opin Microbiol. 2002;5:419–427.
Saito I, Miyamura T, Ohbayashi A, et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549.
Simonetti RG, Cammà C, Fiorello F, et al. Hepatitis C virus infection as a risk factor for hepatocellular carcinoma in patients with cirrhosis. A case-control study. Ann Intern Med. 1992;116:97–102.
Okamoto H, Kurai K, Okada SI, et al. Full length sequence of a hepatitis C virus genome having poor homology to reported isolates—comparative study of four distinct genotypes. Virology. 1992;188:331–341.
Ripoli M, Pazienza V. Impact of hepatitis C virus genetic differences on pathobiology of disease. Expert Rev Anti Infect Ther. 2011;9:747–759.
Moriya K, Fujie H, Shintani Y, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat Med. 1998;4:1065–1717.
Yoshida T, Hanada T, Tokuhisa T, et al. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med. 2002;196:641–653.
Pazienza V, Clement S, Pugnale P, et al. The hepatitis C virus core protein of genotypes 3a and 1b downregulates insulin receptor substrate 1 through genotype-specific mechanisms. Hepatology. 2007;45:1164–1171.
Pazienza V, Clement S, Pugnale P, et al. Gene expression profile of Huh-7 cells expressing hepatitis C virus genotype1b or 3a core proteins. Liver Int. 2009;29:661–669.
Bruno S, Crosignani A, Maisonneuve P, Rossi S, Silini E, Mondelli MU. Hepatitis C virus genotype 1b as a major risk factor associated with hepatocellular carcinoma in patients with cirrhosis: a seventeen-year prospective cohort study. Hepatology. 2007;46:1350–1356.
Raimondi S, Bruno S, Mondelli MU, Maisonneuve P. Hepatitis C virus genotype 1b as a risk factor for hepatocellular carcinoma development: a meta-analysis. J Hepatol. 2009;50:1142–1154.
Levrero M. Viral hepatitis and liver cancer: the case of hepatitis C. Oncogene. 2006;25:3834–3847.
Ripoli M, Barbano R, BalsamoT, Piccoli C, Brunetti V, Coco M, Mazzoccoli G, Vinciguerra M, Pazienza V. Hypermethylated levels of E-cadherin promoter in Huh-7 cells expressing the HCV core protein. Virus Res. 2011;160:74–81.
Takeichi M. Cadherins in cancer: implications for invasion and metastasis. Curr Opin Cell Biol. 1993;5:806–811.
Abid K, Pazienza V, de Gottardi A, et al. An in vitro model of hepatitis C virus genotype 3a-associated triglycerides accumulation. J Hepatol. 2005;42:744–751.
Brooks CL, Gu W. How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer. 2009;9:123–128.
Arora P, Kim EO, Jung JK, Jang KL. Hepatitis C virus core protein downregulates E-cadherin expression via activation of DNA methyltransferase 1 and 3b. Cancer Lett. 2008;261:244–252.
Csepregi A, Röcken C, Hoffmann J, et al. APC promoter methylation and protein expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:579–589.
Dejeux E, Audard V, Cavard C, Gut IG, Terris B, Tost J. Rapid identification of promoter hypermethylation in hepatocellular carcinoma by pyrosequencing of etiologically homogeneous sample pools. J Mol Diagn. 2007;9:510–520.
Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–1064.
Chen J, Zhang B, Wong N, Lo AW, To KF, Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, Yu J, Ng HK, Ling MT, Huang AL, Cai XF, Ko BC. Sirtuin 1 is upregulated in a subset of hepatocellular carcinomas where it is essential for telomere maintenance and tumor cell growth. Cancer Res. 2011;71:4138–149.
Choi HN, Bae JS, Jamiyandorj U, et al. Expression and role of SIRT1 in hepatocellular carcinoma. Oncol Rep. 2011;26:503–510. doi:10.3892/or.2011.1301.
Hp Li, Yw Leu, Ys Chang. Epigenetic changes in virus-associated human cancers. Cell Res. 2005;15:262–271.
Laurson J, Khan S, Chung R, Cross K, Raj K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis. 2010;31:918–926.
Vivekanandan P, Daniel HD, Kannangai R, Martinez-Murillo F, Torbenson M. Hepatitis B virus replication induces methylation of both host and viral DNA. J Virol. 2010;84:4321–4329.
Waris G, Turkson J, Hassanein T, Siddiqui A. Hepatitis C virus (HCV) constitutively activates STAT-3 via oxidative stress: role of STAT-3 in HCV replication. J Virol. 2005;79:1569–1580.
Tacke RS, Tosello-Trampont A, Nguyen V, Mullins DW, Hahn YS. Extracellular hepatitis C virus core protein activates STAT3 in human monocytes/macrophages/dendritic cells via an IL-6 autocrine pathway. J Biol Chem. 2011;286:10847–10855.
Yu H, Jove R. The stats of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–1058.
Turkson J, Kim JS, Zhang S, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–269.
Ulane CM, Rodriguez JJ, Parisien JP, Horvath M. STAT3 ubiquitylation and degradation by mumps virus suppress cytokine and oncogene signaling. J Virol. 2003;77:6385–6393.
Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci USA. 2005;102:6948–6953.
Paredes R, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature Med. 2011;17:330–339.
O’Hagan HM, Mohammad HP, Baylin SB. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155.
Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol. 2009;11:492–500.
Bernier M, Paul RK, Martin-Montalvo A, Scheibye-Knudsen M, Song S, He HJ, Armour SM, Hubbard BP, Bohr VA, Wang L, Zong Y, Sinclair DA, de Cabo R. Negative regulation of STAT3 protein-mediated cellular respiration by SIRT1 protein. J Biol Chem. 2011;286:19270–19279.
Acknowledgments
We thank Prof. Francesco Negro in whose laboratory the plasmids pIRES2-EGFP/Core of different genotypes were generated. This work was supported by the “Italian Ministry of Health” from grant RC1103GA48, through the Research Unit of Gastroenterology, “Casa Sollievo della Sofferenza” IRCCS, San Giovanni Rotondo (FG), Italy.
Conflict of interest
No conflict of interest is declared.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Benegiamo, G., Vinciguerra, M., Mazzoccoli, G. et al. DNA Methyltransferases 1 and 3b Expression in Huh-7 Cells Expressing HCV Core Protein of Different Genotypes. Dig Dis Sci 57, 1598–1603 (2012). https://doi.org/10.1007/s10620-012-2160-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-012-2160-1