Abstract
Plant species that are capable of propagating clonally are expected to be less vulnerable to habitat fragmentation due to their long life span. Cypripedium calceolus L. is a rare, clonal, long-lived orchid species. It has suffered marked decline because of habitat loss and fragmentation and over-collection, yet an IUCN report on this species does not regard fragmentation as a major threat to the species. We applied 13 nuclear microsatellites and cpDNA sequences to identify the patterns of population structure, genetic diversity and connectivity of six remnant local populations of C. calceolus in highly fragmented Gdańsk Pomerania region (N Poland). Despite severe (80%) loss of localities in the studied area we found that the local populations retain high levels of clonal (R 0.86–1) and genetic diversity (He = 0.572). However, their differentiation is relatively high (FST = 0.132 for nuclear SSR and FST = 0.363 for cpDNA) despite close geographic proximity (0.6–57 km). Bayesian clustering classified populations according to their geographic origin with little admixture. Low genetic connectivity between the remnant populations shows that the current gene flow is too low to serve as a cohesive force in a fragmented habitat, which may impede a quick response to environmental change. The species’ ability to retain ancestral variation may help withstand fragmentation, but in the light of observed extirpation rate it should be rather considered as a factor that only delays local populations’ extinction. This leads to the conclusion that habitat loss and fragmentation should be regarded as a real threat to stability of C. calcelolus populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human-induced habitat loss and fragmentation are serious threats to the viability of plant populations that may lead to their decline and extinction (Young et al. 1996; Young and Clarke 2000; Lienert 2004; Reed 2004; Ouborg et al. 2006; Aguilar et al. 2006, 2008; Vranckx et al. 2012). The primary driver of this process is disintegration of large habitats into small, often isolated patches that in consequence leads to overall reduction in population size and numbers. Erosion of genetic variation due to random genetic drift and inbreeding in populations of reduced size compromises their ability to respond to environmental changes and thus increases their risk of extinction (Reed 2004). Sensitivity to this process largely depends on a plant’s life history characteristics (Herault and Honnay 2005; Honnay and Jacquemyn 2007; Lindborg 2007; Lindborg et al. 2012). Plant species that are capable of propagating clonally are expected to be less vulnerable to habitat fragmentation due to their long lifespan (e.g. Honnay and Bossuyt 2005; Bossuyt and Honnay 2006; Van Geert et al. 2008). However, reduced sexual reproduction resulting from environmental changes (e.g., loss of pollinators or genetically compatible mates in fragmented habitats) may lead populations to shift towards a predominately clonal reproductive strategy (Barrett 2015; Lin et al. 2016). Due to a low frequency of seed production and dispersal, clonal plants may be less effective in spreading into new habitat patches and respond more slowly to habitat change than non-clonal species, which may result in local extinction (Buckley and Frecleton 2010; Zobel et al. 2010; Wiberg et al. 2016).
Yellow Lady’s slipper (Cypripedium calceolus L.) is an emblematic, clonal, outcrossing, orchid species that has suffered a marked decrease in the localities and area occupied throughout its Eurasiatic range (Terschuren 1999). Although currently it is regarded as a least concern species according to the IUCN, in some countries it is included in the red list as a threatened species. Fragmentation among its populations at varying level of severity was observed in some countries but this process was not regarded as a major threat to the species (Rankou and Bilz 2014), mainly due to its wide distribution. Results of previous genetic studies on C. calceolus showed relatively high levels of polymorphism and clonal diversity as well as low or moderate levels of population differentiation (Kull and Paaver 1997; Brzosko et al. 2002, 2009, 2011; Gargiulo et al. 2018), which may support the claim that fragmentation has little effect on C. calceolus. However, authors attributed these features to persistence of residual diversity when the species had a more continuous distribution and pointed out that observed population differentiation may also be fragmentation related (Kull and Paaver 1997; Brzosko et al. 2011).
Gene flow, which often acts as a cohesive force in plant population, is rather negligible in C. calceolus due to constraints related to its pollination mechanisms, seed dispersal and germination. Fruit set is relatively low, frequently below 10% (Kull 1998; Nicole et al. 2005; Pedersen et al. 2012; Brzosko et al. 2017b). Although long-distance dispersal of its dust-like seeds is expected to occur (Fay et al. 2009), effective seed dispersal does not exceed 5 m (Brzosko et al. 2017b). C. calceolus also has high specificity for mycorrhizal fungi which are necessary for germination in nature (Shefferson et al. 2005). Seeds germinate at a low rate after long (> 2–3 years) incubation times (Rasmussen and Pedersen 2011). This may make seedling recruitment rates very low, which is supported by usually low (< 10%) percentages of juvenile individuals in populations (Brzosko et al. 2002; Nicole et al. 2005). These characteristics may negatively affect the ability of the species to migrate into new habitat patches in response to climate change and habitat fragmentation (Lindborg et al. 2012; Marini et al. 2012). Thus, the natural extirpation–colonisation dynamic of populations may be negatively affected by fragmentation, which may compromise their stability long before genetic factors become a threat to their persistence.
From nearly 400 localities of this species originally known in Poland, more than half are extinct (Piękoś-Mirkowa and Mirek 2003; Kaźmierczakowa et al. 2014). Locally, the decrease can be even more dramatic. Based on the literature (from 1818 to 2003) and field studies (1993–2014) in Gdańsk Pomerania (northern Poland), we noted the extirpation of 21 out of 24 local populations (Markowski and Buliński 2004; Olszewski and Minasiewicz 2007). Extirpation directly connected with habitat loss accounted for only c. 20%, the rest was due to stochastic events (Olszewski and Minasiewicz unpubl. res.), which may point to the strong indirect influences of habitat fragmentation on the studied species. Detailed genetic study of fine-scale patterns of population connectivity in a highly fragmented landscape is needed to shed more light on the plant response to this increasingly common threat.
Despite the fact that the population genetics of C. calceolus was extensively studied previously, a combination of variable plastid and nuclear markers has never been applied to study populations within such a fine spatial scale and fragmentation context. Nuclear markers can detect both pollen flow and seed dispersal. Maternally inherited chloroplast genome of angiosperms reflects only seed gen flow (Birky 1995). Comparative analysis of both kinds of markers are widely used approaches for obtaining information on genetic structure resulting from different types of gene flow (i.e. pollen vs. seed flow; Ennos 1994; Petit et al. 2005) and may provide valuable information on genetic diversity, and population connectivity of endangered species.
In this study we investigated the genetic diversity and fine-scale patterns of population genetic structure of six, highly fragmented populations of rhizomatous orchid C. calceolus within a narrow geographic area in Gdańsk Pomerania. We applied genetic markers with different modes of inheritance and mutation rates, namely nuclear microsatellite loci (SSR) and plastid DNA (cpDNA). The study addressed the following questions: (1) what is the level of clonal diversity and genetic diversity and their spatial distribution within and among remnant C. calceolus populations in the Gdańsk Pomerania region?; (2) how have historic and contemporary events influenced the current patterns of genetic structure?; (3) what is the relative contribution of seed versus pollen flow to gene flow, and what are the consequences of the observed dispersal patterns for population connectivity in fragmented landscape?
Methods
Study species
Cypripedium calceolus is a perennial orchid distributed mostly in boreal Eurasia (Cribb 1997). It grows in woodlands, rarely in open scrub, predominantly on calcareous soils (Cribb 1997; Kull 1999). Its big, yellow trap flowers offer no rewards to the insect. Plants are pollinated by small or medium-sized bees, but its pollination specificity is rather low (Nilsson 1979; Erneberg and Holm 1999; Antonelli et al. 2009). Like all orchids, it produces a large number of dust-like seeds that are capable of long-distance dispersal (Fay et al. 2009). The species can also reproduce asexually by means of rhizomes. C. calceolus is a long-lived species with longevity ranging from 25 to 100 years, depending on clump size (Kull 1988; Shefferson et al. 2001; Nicole et al. 2005). However, its development till the first flowering may take 10 years, and it is dependent on symbioses with appropriate fungi (Shefferson et al. 2005).
Study site
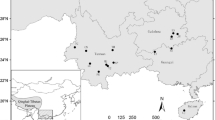
The studies were conducted in Gdańsk Pomerania in northern Poland. C. calceolus can only be found in three main geographic localities in the region, all of which are in fragmented forest patches (Olszewski and Minasiewicz 2007). Because we were interested in fine-scale patterns of genetic structure, we identified spatially isolated (> 0.5 km) remnant patch as separate population. We identified six local populations: three (OL1, OL2, OL3) in Ostrzycki Las Nature Reserve, two (BK, JG) in Dolina Kulawy Nature Reserve and one (PR) in the vicinity of Prokowo village (Fig. 1a, b; Table 1). Both the Ostrzycki Las and Prokowo populations have been mentioned in literature since the end of nineteenth century, although the actual locations of C. calceolus patches in these forest complexes have changed over time (Schultze 1919 after Abromeit et al. 1898–1940; Machnikowski and Fałtynowicz 1982; Olszewski and Minasiewicz, unpubl. res.) suggesting that an extirpation–colonization dynamic of the species is occurring in these locations. Three local populations of the studied species in Ostrzycki Las (average distance 1.5 km) were found in calciphilous beech woods. In 2000, a new patch of C. calceolus was found in a c. 60 year old Picea abies (L.) H. Karst forest in Prokowo, in a place where improvement cutting coupled with ground layer disturbance occurred a few years earlier (Minasiewicz, unpubl. res.). The local populations in Bukówki and Jezioro Głuche were not noted until 1980, in spite of extensive botanical studies conducted in the area (see Izydorek et al. 2008) and presence of suitable habitat. The population in Bukówki (BK) is located on the edge of Carpinion betuli forest type. Its establishment could have coincided with the site disturbance due to construction that led to deposition of lime-rich mud excavate in the surrounding area. It is 2.5 km away from the small population of the study species located on the SE-facing, steep shore of Jezioro Głuche (JG) in a planted Pinus silvestris L. forest with Juniperus communis L. shrubs. Light conditions were relatively good for C. calceolus in all but OL1 and OL2 populations, where plants were overshadowed by adult Fagus sylvatica L.
a Location of Cypripedium calceolus populations in Gdańsk Pomerania. Location of extinct and present populations are marked with a cross and dot, respectively. Pie charts reflect the frequency of occurrence of each chloroplast haplotype in each population. Haplotype colours correspond to those shown in the median-joining network of plastid DNA haplotypes; b The exact locations of C. calceolus patches in Ostrzycki Las (OL) population. c Median-joining network for plastid DNA haplotypes in C. calceolus. The haplotypes are indicated by the circles, the size of each circle being proportional to the observed frequency of each haplotype. The number of mutations required to explain transitions among haplotypes is indicated along the lines connecting the haplotypes by cross hatches
Sample collection and DNA extraction
We sampled C. calceolus in all six extant local populations in the Gdańsk Pomerania region. The distances between populations ranged from 0.6 to 59 km, on average 23 km (Fig. 1a, b). Shoots were selected randomly for sampling within each local population according to Suzuki et al. (2004). Leaf tips (each measuring 2 cm2) of the uppermost leaf were collected and dried in silica gel. A total of 166 shoots were collected, ranging from 14 to 42 samples per local population (Table 1). All necessary permissions were obtained from the relevant authorities. Total genomic DNA was extracted with a CTAB-based method (Bekesiova et al. 1999).
Molecular markers and genotyping assays
Samples of C. calceolus were sequenced for the accD-psal intergenic spacer (IGS) as described by Fay et al. (2009). The sequences were generated on an ABI 3720 automated capillary DNA sequencer. Finch TV (Geospiza) was used to edit the sequences, and the two complementary strands were assembled by AutoAssembler (ABI). All sequences were aligned by eye using SeaView v. 4 (Gouy et al. 2010). Sequences were deposited in GenBank under accession numbers KP902524–KP902531.
Samples of yellow lady’s slipper were also analyzed for diversity in species-specific 13 microsatellite loci (Online Resources S1; Minasiewicz and Znaniecka 2014). All polymerase chain reactions were conducted on an Eppendorf NEXUS GX1 (Eppendorf) following the protocol of Minasiewicz and Znaniecka (2014). Microsatellite genotyping was performed using an ABI 3130 sequencer (Applied Biosystem) and LIZ-500 size standard (Applied Biosystem). Alleles were manually scored and genotypes were determined for each individual using GeneMarker (Soft Genetics LLC).
Data analysis
Genetic diversity of sampled populations
Populations were characterized for diversity based on plastid DNA as gene diversity and the number of haplotypes and haplotype richness using CONTRIB program (Petit et al. 1998) and were corrected for differences in sample size using the rarefaction method.
Nuclear microsatellite diversity of all sampled local populations was calculated as observed (Ho) and expected (He) heterozygosity, number of allele per locus (A), number of effective alleles per locus (Ae) and the number of private alleles, which were calculated in GenAlEx v. 6.5 (Peakall and Smouse 2012). Private allelic richness (PAR) were calculated in HP-RARE (Kalinowski 2005), which uses the rarefaction method (40 genes) to correct for sample size differences. The inbreeding coefficient FIS was calculated in FSTAT (Goudet 2001). Deviations from Hardy–Weinberg were tested in GenAlEx. Linkage disequilibrium (LD) was estimated in GENEPOP 4.2 (Rousset 2008). Standard Bonferroni correction was applied to obtain the proper significance for multiple comparisons (Rice 1989). We used nonparametric Kruskal–Wallis test implemented in STATISTICA v.10 software to test for differences in diversity indices between localities.
SSR viability and clonal diversity assessment
The existence of possible genotyping errors due to stuttering, large allele dropouts, and null alleles was tested using MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004). In clonal plants, multilocus genotypes (MLG) may result from rhizome spread as well as different sexual reproduction events. To investigate this possibility we used the estimator Psex (Arnaud-Haond et al. 2007b). We considered plants with identical multilocus SSR genotypes to originate from vegetative spread if Psex was < 0.05. These duplicate clonal samples were excluded from subsequent analyses of genetic diversity. Clonal diversity was assessed within each population and across all populations by genotypic richness (R Dorken and Eckhert 2001). All calculations were conducted using GENECLONE 2.0 (Arnaud-Haond et al. 2007a).
Population differentiation and structure
The geographical structure of genetic variation in plastid DNA was analysed using a median-joining (MJ) network (Bandelt et al. 1999) based on haplotypes in NETWORK 4.2.0.1. (http://www.fluxus-engineering.com). Ten indels (duplication events) were down weighted in the analysis.
Nuclear genetic differentiation among geographic populations was measured using pairwise FST and RST indices as calculated in ARLEQUIN v. 3.5 (Excoffier and Lischer 2010). The statistical significance was tested using 10,000 permutations. We employed locus-by-locus analysis of molecular variance (AMOVA) to partition within—and among-population genetic variation. Isolation by distance hypotheses of population differentiation were tested based on correlation of log pairwise geographic and genetic differences (linearized genetic distance FST and RST according to Slatkin 1995) based on plastid and nuclear markers using Mantel test (Mantel 1967). All calculations were carried on in the program ARLEQUIN v. 3.5 statistical significance was obtained with 10,000 permutations.
Population structure was also evaluated using Bayesian clustering methods in STRUCTURE 2.3.3 (Pritchard et al. 2010). Ten separate runs of STRUCTURE were performed at each K (the number of putative genetically-defined populations) from 1 to 8 with a burn-in of 250,000 iterations and run-length of 1,000,000 Markov chain Monte Carlo (MCMC) iterations for each run. The admixture model assuming correlated allele frequencies was applied. The optimal K was determined by the Evanno method (DK) (Evanno et al. 2005) and the maximum likelihood method (Pritchard et al. 2000) using STRUCTURE HARVESTER (Earl and von Holdt 2012). CLUMPP software (Jakobsson and Rosenberg 2007) was used to average results for each K value across runs, and summary barplots were built by the program DISTRUCT v.1.1 (Rosenberg 2004). To further understand the clustering patterns, genetic distance-based principal coordinate analysis (PCoA) was carried out using GenAlEx v. 6.5. This multivariate descriptive method is not dependent on any model assumption and can thus provide a useful validation of Bayesian clustering output (Patterson et al. 2006).
Demographic analyses
Genetic signatures of recent bottlenecks were tested using BOTTLENECK 1.2 (Cornuet and Luikart 1996; Piry et al. 1999) with two-phase mutation model (TPM) recommended for microsatellite loci (Di Rienzo et al. 1994). For TPM, 70% of mutations were defined as following a stepwise mutation model running 1000 replications. The significance of the tests was assessed using the Wilcoxon sign-rank test, which is the most appropriate test when fewer than 20 microsatellite loci are used (Piry et al. 1999).
To identify individuals that were immigrants into their sampled location or were of admixed ancestry we run STRUCTURE analysis with K = 4 and prior population information (USEPOPINFO option) and set GENSBACK = 2 (it assesses ancestry up to two generations back) (Pritchard et al. 2010). The number of burn-in iterations, and the total run length were the same as in the previous analysis. Individuals were considered residents if q > 0.8 for the area where they were sampled.
We estimated the ratio of pollen migration (mp) to seed migration (ms) following the protocol of Petit et al. (1992) using the formula:
where FSTc is cytoplasmic variation among sites and FSTn is nuclear variation among sites.
Results
Discrimination of individuals
Samples (166) from six C. calceolus local populations were successfully amplified and examined. Thirteen nuclear microsatellite loci yielded 77 alleles in total. No loci showed the existence of null alleles. The analysis revealed 141 distinct MLGs present in the dataset. The lowest clonal diversity (R) was noted in JG—the smallest local population both in the number of aerial shoots (30) and area occupied. In the remaining populations, clonal diversity was high and ranged from 1 in BK and OL3, where all the sampled plants represented different MLGs, to 0.86 in OL1 (Table 1). The Psex value of putative clones in all the populations studied were below 0.05 and were considered as having originated from vegetative spread, and they were omitted from subsequent analyses.
Plastid and nuclear diversity of sampled populations
Eight haplotypes were identified (Fig. 1c) by the combination of the alleles for the 12 different loci based on variation within accD-psaI IGS (Online Resources Table S2, Table S3) in 62 individuals representing six local populations of C. calceolus. The local populations had from one to three haplotypes (Fig. 1a, b), and they differed in the levels of diversity based on haplotype richness (HR 0.000–1.978) and haplotype diversity. However, when calculations were based on three geographic populations, the level of diversity was similar (HR 2.000–2.712 and HD 0.689–0.741).
All studied populations of C. calceolus exhibited similar, relatively high level of nuclear genetic diversity (Table 2), except JG, in which all sampled shoots represented ramets of the same genetic individual. All loci were polymorphic with mean observed (Ho = 0.587) and expected (He = 0.572) heterozygosity, when JG was excluded from the calculations. Furthermore, populations did not differ significantly in their calculated diversity indices. However, the number of unique alleles and allelic richness after rarefaction differed among populations: OL3 (0.55), BK (0.22), OL2 (0.12) and PR (0.08). All inbreeding coefficients were low and non-significantly different from zero except in JG (Table 2).
Neither the global test for Hardy–Weinberg equilibrium nor population-level tests revealed any deviation from Hardy–Weinberg equilibrium. However, departure from the H-W rule was significant at two loci after Bonferroni correction when all three patches from Ostrzycki Las were merged, but remained non-significant when only OL2 and OL3 were joined, which indicated some substructuring of OL. Several locus pairs showed evidence of linkage disequilibrium in three local populations: in OL1 (Ccal_19–Ccal_25, Ccal_25–Ccal_48, Ccal_25–Ccal_34), OL2 (Ccal_9–Ccal_25, Ccal_25–Ccal_50, Ccal_24–Ccal_48) and PR (Ccal_7–Ccal_24). Since no consistent pairs of loci were linked, all loci were treated as independent because any apparent linkage was probably population-specific (see Frankham et al. 2002; Zartman et al. 2006).
Population differentiation and structure
A haplotype network based on plastid DNA revealed differences among populations. Only three of eight haplotypes (H1, H2, H3) were shared among local populations, with OL sharing three of the four identified haplotypes with other populations. BK had all three unique ones. High genetic differentiation was observed across populations (FST = 0.363). AMOVA results (Table 3) indicate that a high proportion of genetic variability was found among populations (36%, P < 0.0001), whereas within population variation component comprised 64% (P < 0.0001). Pairwise FST comparisons among local populations for cpDNA ranged from 0.000 (OL1/JG) to 1.0 (OL1/Ol2) and were statistically significant (P < 0.001), except for the OL3/PR and OL1/JG pairs (Online Resources Table S4). No signals of isolation by distance (P = 0.8) were detected.
Compared to plastid markers, nuclear markers exhibited lower, but highly significant (P < 0.001) patterns of genetic differentiation (FST = 0.132, RST = 0.127). Similarly to plastid markers, AMOVA analysis revealed that most variance resided within populations at 87% (P < 0.001), whereas 13% (P < 0.001) of the variance was attributed to differences among populations (Table 3). Pairwise comparisons showed that the lowest differentiation was found among the local populations in Ostrzycki Las. Prokowo and Bukówki showed a comparably high level of divergence to Ostrzycki Las as to one another (Online Resources Table S5). All of the applied differentiation metrics were statistically significant. The only exception was RST for the OL2–OL3 pair (P = 0.277).
The analysis of individual multilocus genotypes of the 141 samples using the structure algorithm showed the best clustering solution was at K = 3 (ΔK = 241.4) based on the Evanno method (Online Resources S6) and at K = 4 based on maximum likelihood method [mean Ln P (D) = − 4075.5]. Clustering of individuals for K = 3 clearly corresponded to geographical groups of local populations (Cluster 1 = OL1, OL2, OL3, Cluster 2 = Prokowo, Cluster 3 = Bukówki and Jezioro Głuche). Populations retained the same allocations for K = 4 as for K = 3, except OL1 which was clearly separated from OL2/OL3 cluster (Fig. 2). However, K = 4 provided a better biological explanation than did K = 3, and is concordant with the result of pairwise FST and RST. When OL1 was joined to OL2/OL3, the Wahlund effect also suggested that the OL population could be substructured. PCoA analysis (Fig. 3) generally confirmed the pattern of genetic structure obtained with Bayesian clustering in structure for K = 4. There was also no significant signal of isolation by distance among populations calculated either by FST (P = 0.310) or RST (P = 0.173).
a Geographical representation of the three (K3) and b four (K4) clusters assessed with STRICTURE software. The location, of extinct and present populations of Cypripedium calceolus in Gdańsk Pomerania are marked with a cross and dot, respectively. Pie charts reflect the proportional membership for K = 3 and K = 4 in each population. Colours correspond to those shown in the vertical bar plot; Barplot represent proportional membership (q) of each sampled MLG of Cypripedium calceolus at K = 3 and K = 4, without use of prior population information (USEPOPINFO = 0). Each individual is represented by a horizontal bar, and the length of each bar indicates possibility of membership in each cluster
Detection of recent gene flow and demographic changes
Analysis with STRUCTURE software, in which geographical sampling information for K = 4 was incorporated as a prior, revealed there were no individuals exceeding an established threshold q value (0.800) for any population other than the one in which they were sampled, so no migrants were detected over a time period spanning two generations. Structure identified a total of 25 individuals with q-values ranging from 0.2 to 0.8. These were defined as potentially admixed, because they could not be classified as migrants, but were not clearly assigned as residents either. Estimated pollen migration (mp) among populations was twice as large as estimated seeds migration (ms), (mp/ms = 2).
Bottleneck tests showed evidence of a recent bottleneck in the Ostrzycki Las population (P = 0.045); however, when the local populations were analyzed separately, significant bottlenecks were found for OL1 (P = 0.047) and OL2 (P = 0.027) but not for OL3 (P = 0.684). Additionally, Prokowo exhibited a significant heterozygosity excess under the TPM allele model (P < 0.001).
Discussion
Clonal and genetic diversity of extant populations
Clonal diversity of the populations (R ranged from 0.86 to 1.00) was very high except for the smallest population (JG) where all sampled shoots belonged to the same genetic individual. The populations of Lady’s Slipper Orchid in Gdańsk Pomerania still maintains relatively high levels of genetic diversity as quantified by nuclear genes both in terms of number of alleles (A 3.7–4.8) and heterozygosity (He 0.547–0.600) except for the smallest population JG (A = 1.1; He = 0.038). Similar values for genetic diversity parameters were also reported for Estonian populations (Gargiulo et al. 2018) where the same type of molecular marker (nSSR) was used.
While heterozygosity requires longer periods of fragmentation before its effects become detectable, rare alleles are usually at a greater risk of loss during bottlenecks caused by fragmentation (Young et al. 1996; Kramer et al. 2008). This effect was shown in other long-lived orchid species (Juárez et al. 2011; Chung et al. 2014). Indeed we found that populations with the lowest value of private alleles and private alleles richness (OL1, OL2 and PR) recently passed through a bottleneck. Literature and monitoring data indicate that the events are of a stochastic nature, likely due to a plant decrease in the number due to overshadowing in case of OL1,OL2 and recent founder event in PR.
The level of clonal and genetic diversity revealed in this study may confirm previous observations that genetic diversity of C. calceolus is protected from deterioration by clonal propagation and longevity of individuals and enables long-term stability of the species (Brzosko 2002; Gargiulo et al. 2018). This seems to confirm theoretical prediction by Honnay and Bossuyt (2005) that clonal plants retain genetic variation, which may help them cope better with fragmentation. However, in the light of observed mass extirpation of C. calceolus populations in Gdańsk Pomerania and the extinction debt concept, it should be rather considered as a factor that only delays local populations’ extinction in deteriorated habitat with disrupted connectivity between remnant patches (Hylander and Erhlen 2013). Detailed data on the extirpation–colonisation dynamics of this threatened species from other parts of its range would help to assess the magnitude of the impact that habitat loss and fragmentation may have on the species stability.
Population genetic structure and gene flow in a fragmented landscape
The results of population level F-statistics based on both plastid (FST = 0.363) and nuclear markers (FST = 0.131) as well as Bayesian clustering (Fig. 2) and PCoA (Fig. 3) are concordant and clearly indicate genetic distinctiveness of remnant populations of C. calceolus in Gdańsk Pomerania. The most striking pattern revealed by the use of the plastid marker is relatively strong, among-populations differentiation, which was also found at the level of local populations (patches). Even patches located no more than 3 km apart exhibit substantial differentiation based on cDNA diversity (population OL1 and OL2, 0.6 km apart, FST = 1.00; OL2 and OL3, 1.5 km apart FST = 0.761). Local populations in OL may be remnants of a past, more continuous population in the locality as indicated by the historical distribution of patches (Fig. 1) and the fact that they shared genetic diversity (Fig. 2). However, different plastid types in the local populations suggest that current seed dispersal may be hindered even over short distance (0.6–2.5 km) and/or that plants may encounter infrequent windows of opportunity for successful seed germination and establishment. Patches largely lost their connectivity and are influenced by genetic drift. The two likely oldest populations (OL and PR), as they were noted in the literature from XIX w, located within a range of 15 km, share 40% of their plastid types, which may point to their historic connectivity, although their divergence based on SSRs confirms that current gene flow is insufficient to prevent populations divergence. Also, no seed dispersal was found between Bukówki and the other studied populations in the region, as all haplotypes in studied specimens were unique. Nuclear markers confirm the pattern revealed by plastid markers. Structure revealed no recent (2 generations) migrants; the program assigned the majority of individuals to their populations of origin, classifying non-resident ones as admixed, which is congruent with the hypothesis of past connectivity rather than recent gene flow between populations (Fig. 3). Similar results were obtained in PCoA analysis (Fig. 2).
When genetic structure between genomic compartments was translated into a pollen/seed flow ratio (mp/ms = 2) it revealed that pollen flow is two times more efficient than seed dispersal. Although in orchids and other plants pollen flow may be a strong cohesive force (Pinheiro et al. 2014; Browne and Karubian 2018), in C. calceolus, due to small (rarely longer than 600 m) foraging distance of its putative pollinators (Gathmann and Tscharntke 2002; Zurbuchen et al. 2010), the homogenizing influence of pollen flow on populations’ genetic diversity may be expected only over short distances. Effective seed dispersal distances were even lower in the studied species and did not exceed 5 m (Brzosko et al. 2017b). Although long-distance dispersal of its dust-like seeds is also expected to occur (Fay et al. 2009), constrains connected to its deceptive pollination mechanism (Jersakova et al. 2006) and seed germination (Shefferson et al. 2005) do not make it a very efficient cohesive force. The high rate of population extirpation in the area, coupled with reduced gene flow, may indirectly confirm previous observations that low frequency of seed dispersal and/or establishment in clonal, outcrossing species may impede a quick response to habitat change in a fragmented landscape, which may result in local extinction (Buckley and Frecleton 2010; Zobel et al. 2010).
Habitat fragmentation usually leads to isolation by distance (Zhao et al. 2013), but it was not observed in this study regardless of the marker used. A lack of IBD accompanied by a significant level of genetic differentiation among populations may be caused by overall reduced gene flow or its environmental structuring (Orsini et al. 2013). The latter is rather less possible, as no relation between genetic and environmental (climatic) variation has been observed (Gargiulo et al. 2018). The lack of IBD and significant genetic structure between naturally fragmented Alpine and Tatra populations of C. calceolus (Nicole et al. 2005; Brzosko et al. 2011) are well explained by stochastic colonization and restricted gene flow in spatially isolated mountain regions. A similar pattern was also observed in the lowland Estonian populations (Gargiulo et al. 2018). Hence, reduced gene flow shown in this study, especially when coupled with founder event like the one detected in Prokowo (confirmed by monitoring and genetic data: bottleneck and strong LD), may be responsible for population differentiation.
In Gdańsk Pomerania where about 80% of C. calceolus populations have vanished since the fragmentation process has started, about 120 years ago, the extirpation–colonisation imbalance is clearly visible. The extirpation process of the C. clceolus populations was the most intense during the first 60–70 years from the beginning of the fragmentation process. Then, the most of the local population of this clonal species, known for its longevity, vanished (Świeboda 1976; Olszewski and Minasiewicz 2007). Monitoring did not confirm either recolonization of vacant, suitable habitat or formation of populations in new locations.
This may imply susceptibility of C. calceolus to fragmentation despite clonal and genetic diversity remain largely unaffected by it. The combined use of plastid and nuclear genetic markers revealed that there is low genetic connectivity between the remnant populations. Seed dispersal between them is either too small or ineffective. This leads to a conclusion that changes caused by habitat loss and fragmentation should be considered as a real threat to stability of C. calcelolus populations.
Conservation implications
Bottlenecks and restricted gene flow discovered among the local C. calceolus populations studied as well as the relatively small-sized area occupied by most of them (Table 1) might negatively influence the persistence of these populations in the long term. Therefore, management actions should be taken to prevent further population divergence. First, the management of successional processes (canopy closure) should be planned. Reintroduction to locations from which C. calceolus has vanished because of natural processes, but where environmental conditions remain favorable (Olszewski and Minasiewicz, unpubl. res), would also be advisable to reinforce gene flow among populations and to slow divergence processes. Supplemental pollination could also enhance fruiting success (Ramsay and Stuart 1998; Brzosko et al. 2017a) and the frequency of seed dispersal.
References
Abromeit J, Neuhof W, Steffen H (1898–1940) Flora von Ost-und Westpreussens. 1(1898), 2(1903), II (1926), III (1931), IV (1934), Schlussband (1940) R. Friedlander u. Sohn, Berlin. Preuss. Bot. Verl. E.V., Konigsberg
Aguilar R, Ashworth L, Gaaletto L, Azien MA (2006) Plant reproductive susceptibility to habitat fragmentation: review and synthesis through a meta-analysis. Ecol Lett 9:968–980
Aguilar R, Quesada M, Ashworth L, Herrerias-Diego Y, Lobo J (2008) Genetic consequences of habitat fragmentation in plants populations: susceptible signals in plant traits and methodological approaches. Mol Ecol 17:5177–5188
Antonelli A, Dahlberg JC, Carlgren KHI, Appleqvist T (2009) Pollination of the lady’s slipper orchid (Cypripedium calceolus) in Scandinavia. Nordic J Bot 27:1–8
Arnaud-Haond S, Belkhir K (2007a) GENCLONE: a computer program to analyze genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7:15–17
Arnaud-Haond S, Duarte CM, Alberto F, Serrão EA (2007b) Standardizing methods to address clonality in population studies. Mol Ecol 16:5115–5513
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 1:37–48
Barrett SCH (2015) Influences of clonality on plant sexual reproduction. Proc Natl Acad Sci 112(29):8859–8866. https://doi.org/10.1073/pnas.1501712112
Bekesiova I, Nap JP, Mlynarova L (1999) Isolation of high quality DNA from leaves of carnivorous plant Drosera rotundifolia. Plant Mol Biol Rep 17:269–277
Birky CW (1995) Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc Natl Acad Sci USA 92:11331–11338
Bossuyt B, Honnay O (2006) Interactions between plant life span, seed dispersal capacity and fecundity determine metapopulation viability in a dynamic landscape. Landscape Ecol 21(8):1195–1205. https://doi.org/10.1007/s10980-006-0016-9
Browne L, Karubian J (2018) Habitat loss and fragmentation reduce effective gene flow by disrupting seed dispersal in a neotropical palm. Mol Ecol 27(15):3055–3069. https://doi.org/10.1111/mec.14765
Brzosko E (2002) Dynamics of island populations of Cypripedium calceolus in the Biebrza river valley (NE Poland). Bot J Linn Soc 139:67–77
Brzosko E, Wróblewska A, Ratkiewicz M (2002) Spatial genetic structure and clonal diversity of Island population of Lady’s slipper (Cypripedium calceolus) from the Biebrza National Park (northeast Poland). Mol Ecol 11:2499–2509
Brzosko E, Wróblewska A, Ratkiewicz M, Till-Bottraud I, Nicole F, Baranowska U (2009) Resources of Cypripedium calceolus genetic diversity at the edge and in the centre of European geographical range. Ann Bot Fenn 46:201–214
Brzosko E, Wróblewska A, Tałałaj I, Wasilewska E (2011) Genetic diversity of Cypripedium calceolus in Poland. Plant Syst Evol 295:83–96
Brzosko E, Ostrowiecka B, Mirski P, Jermakowicz E, Tałałaj I, Wróblewska A (2017a) Pollinator limitation affects low reproductive success in populations of nectarless orchid in the Biebrza National Park. Acta Agrobot 70(1):1706. https://doi.org/10.5586/aa.1706
Brzosko E, Ostrowiecka B, Kotowicz J, Bolesta M, Gromotowicz A, Gromotowicz M, Orzechowska A, Orzołek J, Wojdalska M (2017b) Seed dispersal in six species of terrestrial orchids in Biebrza National Park (NE Poland). Acta Soc Bot Poloniae. https://doi.org/10.5586/asbp.3557
Buckley HI, Frecleton RP (2010) Understanding the role of species dynamics in abundance–occupancy relationships. J Ecol 98:645–658
Chung MY, Nason JD, López-Pujol J, Yamashiro T, Yang BY, Luo YB, Chung MG (2014) Genetic consequences of fragmentation on populations of the terrestrial orchid Cymbidium goeringii. Biol Conserv 170:222–231
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cribb P (1997) The genus Cypripedium. Timber Press, Inc. Portland, Oregon
Di Rienzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Dorken ME, Eckhert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol 89:339–350
Earl D, von Holdt ED (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Gen Res 4:359–361
Ennos RA (1994) Estimating the relative rates of pollen and seed migration among plant populations. Heredity 72:250–259
Erneberg M, Holm B (1999) Bee size and pollen transfer in Cypripedium calceolus (Orchidaceae). Nordic J Bot 19:363–367
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Fay MF, Bone R, Cook P, Kahandawala I, Greensmith J, Harris S, Pedersen H, Ingrouille MJ, Lexer Ch (2009) Genetic diversity in Cypripedium calceolus (Orchidaceae) with a focus on north-western Europe, as revealed by plastid DNA length polymorphisms. Ann Bot 104:517–525
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
Gargiulo R, Ilves A, Kaart T, Fay MF, Kull T (2018) High genetic diversity in a threatened clonal species, Cypripedium calceolus (Orchidaceae), enables long-term stability of the species in different biogeographical regions in Estonia. Bot J Linn Soc 186(4):560–571. https://doi.org/10.1093/botlinnean/box105
Gathmann A, Tscharntke T (2002) Foraging ranges of solitary bees. J Anim Ecol 71:757–764
Goudet J (2001) FSTAT Ver. 2.9.3, a program to estimate and test gene diversities and fixation indices. http://www.unil.ch/izea/softwares/fstat.html
Gouy M, Guindon S, Gascuel O (2010) SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224
Herault B, Honnay O (2005) The relative importance of local, regional and historical factors determining the distribution of plants in fragmented riverine forests: an emergent group approach. J Biogeogr 32:2069–2208
Honnay O, Bossuyt B (2005) Prolonged clonal growth: escape route or route to extinction? Oikos 10:427–432
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plant species to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
Hylander K, Ehrlén J (2013) The mechanisms causing extinction debts. Tree Ecol Evol 28(6):341–346
Izydorek I, Szadkowska-Izydorek M, Minasiewicz J (2008) Nowe stanowisko Cypripedium calceolus (Cypripediaceae) na Pomorzu Gdańskim [A new locality of Cypripedium calceolus (Cypripediaceae) in the Gdańsk Pomerania]. Fragm Flor Geobot 15:323
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jersáková J, Johnson SD, Kindlmann P (2006) Mechanisms and evolution of deceptive pollination in orchids. Biol Rev 81(2):219–235
Juárez L, Montaña C, Ferrer MM (2011) Genetic structure at patch level of the terrestrial orchid Cyclopogon luteoalbus (Orchidaceae) in a fragmented cloud forest. Plant Syst Evol 297:237–251
Kalinowski ST (2005) HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Kaźmierczakowa R, Zarzycki K, Mirek Z (2014) Polska czerwona księga roślin. Paprotniki i rośliny kwiatowe. Polish Red Data Book of Plants. Wyd. III uaktualnione i rozszerzone. Ss, 895. Instytut Ochrony Przyrody PAN, Kraków
Kramer AT, Ison JL, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics. Conserv Biol 22:878–885
Kull T (1988) Identification of clones in Cypripedium calceolus (Orchidaceae). Proc. Estonian Acad Sci Biol Ecol 37:195–199
Kull T (1998) Fruit-set and recruitment in populations of Cypripedium calceolus L. in Estonia. Bot J Linn Soc 126(1–2):27–38
Kull T (1999) Cypripedium calceolus L. Biological flora of The British Isles. J Ecol 87:913–924
Kull T, Paaver T (1997) Patterns of aspartate aminotransferase variation in relation to population size, founder effect and phytogeographic history in Cypripedium calceolus L. Proc. Estonian Acad Sci Biol Ecol 46:4–11
Lienert J (2004) Habitat fragmentation effects on fitness of plant populations—a review. J Nat Conserv 12:53–72
Lin C, Miriti MN, Goodell K (2016) Demographic consequences of greater clonal than sexual reproduction in Dicentra canadensis. Ecol Evol 6(12):3871–3883. https://doi.org/10.1002/ece3.2163
Lindborg R (2007) Evaluating the distribution of plant life-history traits in relation to current and historical landscape configurations. J Ecol 95:555–564
Lindborg R, Helm A, Bommarco R, Heikkinen RK, Kühn I, Pykälä J, Pärtel M (2012) Effect of habitat area and isolation on plant trait distribution in European forests and grasslands. Ecography 35:356–363
Machnikowski M, Fałtynowicz W (1982) Interesujące gatunki roślin naczyniowych rezerwatu”Ostrzycki Las” na Pojezierzu Kaszubskim. Zeszyty Naukowe Wydziału Biologii I Nauk o Ziemi Uniwersytetu Gdańskiego. Biologia 3:55–62
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Marini L, Bruun HH, Heikkinen RK, Helm A, Honnay O, Krauss J, Kühn I, Lindborg R et al (2012) Traits related to species persistence and dispersal explain changes in plant communities subjected to habitat loss. Divers Distrib 18:898–908
Markowski R, Buliński M (2004) Ginące i zagrożone rośliny naczyniowe Pomorza Gdańskiego. Acta Bot Cass, Monographie 1
Minasiewicz J, Znaniecka JM (2014) Characterization of 15 novel microsatellite loci for Cypripedium calceolus (Orchidaceae) using MiSeq sequencing. Conserv Gen Res 6:527–529
Nicole F, Brzosko E, Till-Bottraud I (2005) Population viability analysis of Cypripedium calceolus in a protected area: longevity, stability and persistence. J Ecol 93:716–726
Nilsson LA (1979) Anthecological studies on the lady’s slipper, Cypripedium calceolus (Orchidaceae). Bot Notiser 132:329–347
Olszewski TS, Minasiewicz J (2007) Cypripedium calceolus na Pomorzu Gdańskim. Botanika w Polsce, sukcesy, problem, perspektywy. 54 Zjazd PTB, Szczecin. Streszczenia referatów i posterów
Orsini L, Vanoverbeke J, Swillen I, Mergeay J, Meester L (2013) Drivers of population genetic differentiation in the wild: isolation by dispersal limitation, isolation by adaptation and isolation by colonization. Mol Ecol 22:5983–5999
Ouborg NJ, Vergeer P, Mix C (2006) The rough edges of the conservation genetics paradigm for plants. J Ecol 94:1233–1248
Patterson N, Price AL, Reich D (2006) Population structure and Eigen analysis. PLoS Genet 2:e190. https://doi.org/10.1371/journal.pgen.0020190
Peakall R, Smouse PE (2012) Genalex 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539
Pedersen H. Rasmussen HN, Kahandawala IM, Fay MF (2012) Genetic diversity, compatibility patterns and seed quality in isolated populations of Cypripedium calceolus (Orchidaceae). Conserv Gen 13:89–98. https://doi.org/10.1007/s10592-011-0267-0
Petit RJ, Kremer A, Bacilieri R, Ducousso A, Zanetto A (1992) Structuration genetique chez les chenes blancs europeens. Complexes d’espe`ces, flux de ge`nes et ressources ge´ne´tiques des plantes. Bureau des Ressources Ge´ne´tiques, Paris, Lavoisier, Cachan, 155–163
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Cons Biol 12:844–855
Petit RJ, Duminil J, Fineschi S, Hampe A, Salvini D, Vendramin GG (2005) Comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol Ecol 14:689 – 701
Piękoś-Mirkowa H, Mirek Z (2003) Flora Polski. Atlas roślin chronionych. Multico Oficyna Wydawnicza, Warszawa
Pinheiro F, Cozzolino S, Draper D, de Barros F, Felix LP, Fay MF, Palma-Silva C (2014) Rock outcrop orchids reveal the genetic connectivity and diversity of inselbergs of northeastern Brazil. BMC Evol Biol 14:49. https://doi.org/10.1186/1471-2148-14-49
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a program for detecting recent effective population size reductions from allele data frequencies. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:954–959
Pritchard JK, Wen W, Falush D (2010) Documentation for STRUCTURE software: version 2.3. http://pritch.bsd.uchicago.edu/structure.html
Ramsay MM, Stewart J (1998) Re-establishment of the lady’s slipper orchid (Cypripedium calceolus L.) in Britain. Bot J Linn Soc 126:173–181. https://doi.org/10.1111/j.1095-8339.1998.tb02524.x
Rankou H, Bilz M (2014) Cypripedium calceolus. The IUCN red list of threatened species 2014: e.T162021A43316125. http://dx.doi.org/10.2305/IUCN.UK.2014-1.RLTS.T162021A43316125
Rasmussen HN, Pedersen H (2011) Cypripedium calceolus germination in situ: seed longevity, and dormancy breakage by long incubation and cold winters. Eur J Environ Sci 1(2):69–70
Reed DH (2004) Extinction risk in fragmented habitat. Animal Conserv 7:181–191
Rice WR (1989) Analysing tables of statistical tests. Evolution 43:223–225
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Shefferson RP, Sandercock BK, Proper J, Beissinger SR (2001) Estimating dormancy and survival of a rare herbaceous perennial using mark-recapture models. Ecology 82:145–156
Shefferson RP, Weiss M, Kull T, Taylor DL (2005) High specificity generally characterizes mycorrhizal association in rare lady’s slipper orchids, genus Cypripedium. Mol Ecol 14:613–626
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Suzuki JI, Herben T, Maki M (2004) An under-appreciated difficulty: sampling of plant populations for analysis using molecular markers. Evol Ecol 18:625–646
Świeboda M (1976) Rozmieszczenie obuwika pospolitego Cypripedium calceolus L. w Polsce. The geographical distribution of Lady’s Slipper Cypripedium calceolus L. in Poland. Ochr Przyr 41:205–230
Terschuren J (1999) Action plan for Cypripedium calceolus in Europe. Nature and environment, no. 100. Council of Europe
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Van Geert A, Fabienne Van Rossum Æ, Triest ÆL (2008) Genetic diversity in adult and seedling populations of Primula vulgaris in a fragmented agricultural landscape. Conserv Genet 9:845–853
Vranckx GUY, Jacquemyn H, Muys B, Honnay O (2012) Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv Biol 26:228–237
Wiberg R, Axel W, Scobie AR, A’Hara SW, Ennos R, Cottrell J (2016) The genetic consequences of long term habitat fragmentation on a self-incompatible clonal plant, Linnaea borealis L. Biol Conserv 201:405–413. https://doi.org/10.1016/j.biocon.2016.07.032
Young AG, Clarke GM (2000) Genetics, demography and viability of fragmented populations. Cambridge University Press, Cambridge
Young AG, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Tree Ecol Evol 11:413–419
Zartman C, McDaniel SF, Shaw AJ (2006) Experimental habitat fragmentation increases linkage disequilibrium but does not affect genetic diversity or population structure in the Amazonian liverworth Radula flaccida. Mol Ecol 15:2305–2315
Zhao Y, Vrieling K, Liao H, Xiao M, Zhu Y, Rong J, Zhang W, Wang Y, Yang J, Chen J, Song Z (2013) Are habitat fragmentation, local adaptation and isolation-by-distance driving population divergence in wild rice Oryza rufipogon. Mol Ecol 22:5531–5547
Zobel M, Moora M, Herben T (2010) Clonal mobility and its implications for spatio-temporal patterns of plant communities: what do we need to know next. Oikos 119:802–806
Zurbuchen A, Landert L, Klaiber J, Müller A, Hein S, Dorn S (2010) Maximum foraging ranges in solitary bees: only few individuals have the capability to cover long foraging distances. Biol Conserv 143:669–676
Acknowledgements
We thank dr. Tomasz S. Olszewski for helping in field work and useful suggestions. Two anonymous reviewers for valuable comments on the manuscript. This study was funded by the Polish National Science Centre Grant No. 3984/B/PO1/2010/39.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Minasiewicz, J., Znaniecka, J.M., Górniak, M. et al. Spatial genetic structure of an endangered orchid Cypripedium calceolus (Orchidaceae) at a regional scale: limited gene flow in a fragmented landscape. Conserv Genet 19, 1449–1460 (2018). https://doi.org/10.1007/s10592-018-1113-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-018-1113-4