Abstract
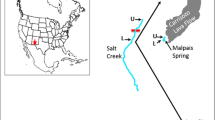
The use of genetic methods to quantify the effects of anthropogenic habitat fragmentation on population structure has become increasingly common. However, in today’s highly fragmented habitats, researchers have sometimes concluded that populations are currently genetically isolated due to habitat fragmentation without testing the possibility that populations were genetically isolated before European settlement. Etheostoma raneyi is a benthic headwater fish restricted to river drainages in northern Mississippi, USA, that has a suite of adaptive traits that correlate with poor dispersal ability. Aquatic habitat within this area has been extensively modified, primarily by flood-control projects, and populations in headwater streams have possibly become genetically isolated from one another. We used microsatellite markers to quantify genetic structure as well as contemporary and historical gene flow across the range of the species. Results indicated that genetically distinct populations exist in each headwater stream analyzed, current gene flow rates are lower than historical rates, most genetic variation is partitioned among populations, and populations in the Yocona River drainage show lower levels of genetic diversity than populations in the Tallahatchie River drainage and other Etheostoma species. All populations have negative FIS scores, of which roughly half are significant relative to Hardy–Weinberg expectations, perhaps due to small population sizes. We conclude that anthropogenic habitat alteration and fragmentation has had a profoundly negative impact on the species by isolating E. raneyi within headwater stream reaches. Further research is needed to inform conservation strategies, but populations in the Yocona River drainage are in dire need of management action. Carefully planned human-mediated dispersal and habitat restoration should be explored as management options across the range of the species.



Similar content being viewed by others
References
Adams SB, Warren ML Jr, Haag WR (2004) Spatial and temporal patterns of fish assemblages of upper coastal plain streams, Mississippi, USA. Hydrobiology 528:45–61
Beerli P (2010) MIGRATE version 3.2: a maximum likelihood and Bayesian estimator of gene flow using the coalescent. http://popgen.scs.edu/migrate.html. Accessed 5 December 2010
Beerli P, Felsenstein J (1999) Maximum likelihood estimation of migration rates and population numbers of two populations using a coalescent approach. Genetics 152:763–773
Beneteau CL, Mandrak NE, Heath DD (2007) Characterization of eight polymorphic microsatellite DNA markers for the greenside darter, Etheostoma blennioides (Percidae). Mol Ecol Notes 7:641–643
Beneteau CL, Mandrak NE, Heath DD (2009) The effects of river barriers and range expansion of the population genetic structure and stability in greenside darter (Etheostoma blennioides) populations. Conserv Genet 10:477–487
Benjamini Y, Yekutieli D (2001) The control of false discovery rate under dependency. Ann Stat 29:1165–1188
Bensch S, Andren H, Hansson B, Pedersen HC, Sand H, Sejberg D, Wabakken P, Akesson M, Liberg O (2006) Selection for heterozygosity gives hope to a wild population of inbred wolves. PLoS ONE. doi:10.1371/journal.pone.0000072
Bohonak AJ (2002) IBD (isolation by distance): a program for analyses of isolation by distance. J Hered 93:153–154
Chapman SS, Griffith GE, Omernik JM, Comstock JA, Beiser MC, Johnson D (2004) Ecoregions of Mississippi, (color poster with map, descriptive text, summary tables, and photographs): Reston, Virginia, U.S. Geological Survey (map scale 1:1,000,000). http://www.epa.gov/wed/pages/ecoregions/ms_eco.htm. Accessed 20 February 2011
Chiucchi JE, Gibbs HL (2010) Similarity of contemporary and historical gene flow among highly fragmented populations of and endangered rattlesnake. Mol Ecol 19:5345–5358
Cooper CM, Knight SS (1991) Water quality cycles in two hill land streams subjected to natural, municipal, and non-point agricultural stresses in the Yazoo Basin of Mississippi, USA (1985–1987). Organ Int Ver Theor Angew Limnol 24:1654–1663
Edberg KL (2009) The effects of a reservoir on genetic isolation in two species of darters. Thesis, Western Kentucky University
Estes-Zumpf WA, Rachlow JL, Waits LP, Warheit KI (2010) Dispersal, gene flow, and population genetic structure in the pygmy rabbit (Brachylagus idahoensis). J Mammal 91:208–219
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetic analysis. Evol Bioinform Online 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure: extensions to linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fluker BL, Kuhajda BR, Lang NJ, Harris PM (2010) Low genetic diversity and small long-term population sizes in the spring endemic watercress darter, Etheostoma nuchale. Conserv Genet 11:2267–2279
Gabel JM, Dakin EE, Freeman BJ, Porter BA (2008) Isolation and identification of eight microsatellite loci in the Cherokee darter (Etheostoma scotti) and their variability in other members of the genera Etheostoma, Ammocrypta, and Percina. Mol Ecol Res 8:149–151
George AL, Caldieraro JB, Chartrand KM, Mayden RL (2008) Population genetics of the blue shiner, Cyprinella caerulea. Southeast Nat 7:637–650
Goudet J (2001) FSTAT version 2.9.3, a program to estimate and test gene diversities and fixation indices. http//:unil.ch/izea/softwares/fstat.html. Accessed 5 December 2010
Guillot G, Santos F (2009) A computer program to simulate multilocus genotype data with spatially auto-correlated allele frequencies. Mol Ecol Res 9:1112–1120
Guillot G, Estoup A, Mortier F, Cosson J-F (2005) A spatial model for landscape genetics. Genetics 170:1261–1280
Hallerman E, Brown B, Epifanio J (2003) An overview of classical and molecular genetics. In: Hallerman EM (ed) Population genetics: principles and applications for fisheries scientists. American Fisheries Society, Bethesda, pp 3–20
Haponski AE, Bollin TL, Jedlicka MA, Stepien CA (2009) Landscape genetic patterns of the rainbow darter Etheostoma caeruleum: a catchment analysis of mitochondrial DNA sequences and nuclear microsatellites. J Fish Biol 75:2244–2268
Hubisz M, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9:1322–1332
Hudman SP, Grose MJ, Landis JB, Skalski GT, Wiley EO (2008) Twenty-three microsatellite DNA loci for population genetic studies and parentage assignment in orangethroat darter, Etheostoma spectabile. Mol Ecol Res 8:1483–1485
Jelks HL, Walsh SJ, Burkhead NM, Contreras-Balderas S, Diaz-Pardo E, Hendrickson DA, Lyons J, Mandrak NE, McCormick F, Nelson JS, Plantania SP, Porter BA, Renaud CB, Schmitter-Soto JJ, Taylor EB, Warren ML Jr (2008) Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries 33:372–407
Jensen JL, Bohonak AJ, Kelley ST (2005) Isolation by distance, web service. BMC Genet 6:13. Version 3.16. http://ibdws.sdsu.edu/. Accessed 1 March 2011
Johnson RL (2007) Geneflow and genetic structuring of yellow cheek darters (Etheostoma moorei) in the little Red River watershed. Final report submitted to the Arkansas Game and Fish Commission, Little Rock, AR
Johnston CE, Haag WR (1996) Life history of the Yazoo darter (Percidae: Etheostoma raneyi), a species endemic to north-central Mississippi. Tulane Stud Zool Bot 30:47–60
Khudamrongsawat J, Heath LS, Heath HE, Harris PM (2007) Microsatellite DNA primers for the endangered vermilion darter, Etheostoma chermocki, and cross-species amplification in other darters (Percidae: Etheostoma). Mol Ecol Notes 7:811–813
Kuhajda BR, George AL, Williams JD (eds) (2008) The desperate dozen: southeastern freshwater fishes on the brink. Southeast Fish Counc Proc 51:10–30
Lamphere BA, Blum MJ (2012) Genetic estimates of population structure and dispersal in a benthic stream fish. Ecol Fresh Fish 21:75–86
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Narum S (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv Genet 7:783–787
NatureServe (2011) http://www.natureserve.org/explorer/servlet/NatureServe?sourceTemplate=tabular_report.wmt&loadTemplate=species_RptComprehensive.wmt&selectedReport=RptComprehensive.wmt&summaryView=tabular_report.wmt&elKey=100184&paging=home&save=true&startIndex=1&nextStartIndex=1&reset=false&offPageSelectedElKey=100184&offPageSelectedElType=species&offPageYesNo=true&post_processes=&radiobutton=radiobutton&selectedIndexes=100184. Accessed 19 February 2011
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Powers SL, Warren ML Jr (2009) Phylogeography of three snubnose darters (Percidae: Subgenus Ulocentra) endemic to the southeastern U.S. Coastal Plain. Copeia 2009:523–528
Powers SL, Mayden RL, Etnier DA (2004) Conservation genetics of the ashy darter, Etheostoma cinereum (Percidae: subgenus Allohistium), in the Cumberland and Tennessee Rivers of the southeastern United States. Copeia 2004:632–637
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pudovkin AI, Zaykin DV, Hedgecock D (1996) On the potential for estimating the effective number of breeders from heterozygosity excess in progeny. Genetics 144:383–387
Rasmussen DI (1979) Sibling clusters and genotypic frequencies. Am Nat 113:948–951
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reed DH (2007) Natural selection and genetic diversity. Heredity 99:1–2
Reed DH (2008) Effects of population size on population viability: from mutation to environmental catastrophes. In: Carroll SP, Fox CW (eds) Conservation biology. Oxford University Press, New York, pp 16–34
Reed DH, Teoh VH, Stratton GE, Hataway JA (2011) Levels of gene flow among populations of a wolf spider in a fragmented habitat: current versus historical rates. Conserv Genet 12:331–335
Rousset F (2008) GENEPOP ‘007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Res 8:103–106
Shields FD, Knight SS, Cooper CM (1994) Effects of channel incision on base flow stream habitats and fishes. Environ Manag 18:43–57
Shields FD, Knight SS, Cooper CM (1998) Rehabilitation of aquatic habitats in warmwater streams damaged by channel incision in Mississippi. Hydrobiology 382:63–86
Simon A, Darby SE (1997) Disturbance, channel evolution and erosion rates: Hotophia Creek, Mississippi. In: Wang SSY, Langendoen EJ, Shields FD Jr (eds) Management of landscapes disturbed by channel incision: stabilization, rehabilitation, restoration. University of Mississippi Press, Oxford, pp 476–481
Skalski GT, Landis JB, Grose MJ, Hudman SP (2008) Genetic structure of creek chub, a headwater minnow, in an impounded river system. Trans Am Fish Soc 137:962–975
Spiegelhalter DJ (2002) Bayesian measures of model complexity and fit. J R Stat Soc Ser B (Methodol) 64:583–639
Sterling K, Warren ML Jr, Noonan BP, Reed DH, Henderson LG (2011) Yazoo darter, Etheostoma raneyi: population and demographic status, distributional changes, and habitat use of an endemic, nongame species. Final report submitted to the Mississippi Museum of Natural Science, Jackson MS
Storz JF (1999) Genetic consequences of mammalian social structure. J Mammal 80:553–569
Storz JF, Bhat HR, Kunz TH (2001) Genetic consequences of polygyny and social structure in an Indian fruit bat, Cynopterus sphinx. I. Inbreeding, outbreeding, and population subdivision. Evolution 55:1215–1223
Suttkus RD, Bailey RM, Bart HL Jr (1994) Three new species of Etheostoma, subgenus Ulocentra, from the Gulf Coastal Plain of southeastern United States. Tulane Stud Zool Bot 29:97–126
Switzer JF, Welsh SA, King TL (2008) Microsatellite DNA primers for the candy darter, Etheostoma osburni, and variegate darter, Etheostoma variatum, and cross-species amplification in other darters (Percidae). Mol Ecol Res 8:335–338
Taggart JB, Haynes RA, Prodohl PA, Ferguson A (1992) A simplified protocol for routine total DNA isolation from salmonid fishes. J Fish Biol 40:963–965
Thompson KW, Muncy RJ (1986) Darters of the Little Tallahatchie watershed in northern Mississippi. J Miss Acad Sci 31:63–77
Tonnis BD (2006) Microsatellite DNA markers for the rainbow darter, Etheostoma caeruleum (Percidae), and their potential utility for other darter species. Mol Ecol Notes 6:230–232
Turner TF, Robison HW (2006) Genetic diversity of the Caddo madtom, Noturus taylori, with comments on factors that promote genetic divergence in fishes endemic to the Ouachita highlands. Southwest Nat 51:338–345
Turner TF, Trexler JC (1998) Ecological and historical associations of gene flow in darters (Teleostei: Percidae). Evolution 52:1781–1801
Warren ML Jr, Burr BM, Walsh SJ, Bart HL Jr, Cashner RC, Etnier DA, Freeman BJ, Kuhajda BR, Mayden RL, Robison HW, Ross ST, Starnes WC (2000) Diversity, distribution, and conservation status of the native freshwater fishes of the southern United States. Fisheries 25:7–29
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Acknowledgments
We thank S. Adams and W. Haag for sharing their experience and advice, M. Bland, A. Commens-Carson, G. McWhirter, B. Sterling, and W. Sterling for their dedicated help in the field collecting samples, C. Jenkins for logistical support, and G. Henderson who produced Fig. 1. We also thank A. Comeault, D. Hataway and S. Nielsen for their assistance in the laboratory and S. Bingham at the Arizona State University DNA Lab. Special thanks to D. Drennen, M. Roberts, and T. Slack for their cooperation and support. This work was supported by funds from the Mississippi Museum of Natural Science, the Mississippi Ecological Services Office, U.S. Fish and Wildlife Service, and the Center for Bottomland Hardwoods Research, Southern Research Station, USDA Forest Service.
Author information
Authors and Affiliations
Corresponding author
Additional information
D. H. Reed—Deceased.
Rights and permissions
About this article
Cite this article
Sterling, K.A., Reed, D.H., Noonan, B.P. et al. Genetic effects of habitat fragmentation and population isolation on Etheostoma raneyi (Percidae). Conserv Genet 13, 859–872 (2012). https://doi.org/10.1007/s10592-012-0335-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0335-0